Comparison of microbiome community structure and dynamics during anaerobic digestion of different renewable solid wastes
- PMID: 40255248
- PMCID: PMC12008556
- DOI: 10.1016/j.crmicr.2025.100383
Comparison of microbiome community structure and dynamics during anaerobic digestion of different renewable solid wastes
Abstract
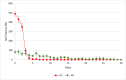
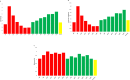
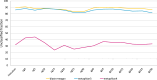
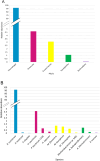
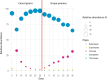
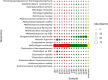
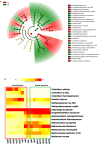
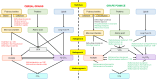
This study analysed the effect of the different lignocellulose composition of two crop substrates on the structure and dynamics of bacterial communities during anaerobic digestion (AD) processes for biogas production. To this end, cereal grains and grape pomace biomasses were analysed in parallel in an experimental AD bench-scale system to define and compare their metagenomic profiles for different experimental time intervals. The bacterial community structure and dynamics during the AD process were detected and characterised using high-resolution whole metagenomic shotgun analyses. Statistical evaluation identified 15 strains as specific to two substrates. Some strains, like Clostridium isatidis, Methanothermobacter wolfeii, and Methanobacter sp. MB1 in cereal grains, and Acetomicrobium hydrogeniformans and Acetomicrobium thermoterrenum in grape pomace, were never before detected in biogas reactors. The presence of bacteria such as Acetomicrobium sp. and Petrimonas mucosa, which degrade lipids and protein-rich substrates, along with Methanosarcina sp. and Peptococcaceae bacterium 1109, which tolerate high hydrogen pressures and ammonia concentrations, suggests a complex syntrophic community in lignin-cellulose-enriched substrates. This finding could help develop new strategies for the production of a tailor-made microbial consortium to be inoculated from the beginning of the digestion process of specific lignocellulosic biomass.
Keywords: Anaerobic digestion; Biogas production; Lignin biodegradation; Lignocellulosic biomasses; Metagenomics.
© 2025 The Authors. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Agbonifo P.E. Renewable energy development: opportunities and barriers within the context of global energy politics. Int. J. Energy Econ. Policy. 2021;11:141–148. doi: 10.32479/ijeep.10773. - DOI
-
- Agregán R., Lorenzo J.M., Kumar M., Shariati M.A., Khan M.U., Sarwar A., Sultan M., Rebezov M., Usman M. Anaerobic digestion of lignocellulose components: challenges and novel approaches. Energies. 2022;15:8413. doi: 10.3390/en15228413. - DOI
-
- Ahuja V., Sharma C., Paul D., Dasgupta D., Saratale G.D., Banu J.R., Yang Y., Bhatia S.K. Unlocking the power of synergy: cosubstrate and coculture fermentation for enhanced biomethane production. Biomass Bioenergy. 2024;180 doi: 10.1016/j.biombioe.2023.106996. - DOI
-
- Akyol Ç., Ince O., Bozan M., Ozbayram E.G., Ince B. Biological pretreatment with Trametes versicolor to enhance methane production from lignocellulosic biomass: a metagenomic approach. Ind. Crops Prod. 2019;140 doi: 10.1016/j.indcrop.2019.111659. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

