Engineering Cell-Specific Protein Delivery Vehicles for Erythroid Lineage Cells
- PMID: 40255284
- PMCID: PMC12006860
- DOI: 10.1021/acsbiomedchemau.4c00098
Engineering Cell-Specific Protein Delivery Vehicles for Erythroid Lineage Cells
Abstract
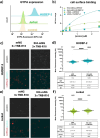
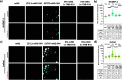
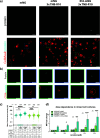
Biologics such as proteins, peptides, and oligonucleotides are powerful ligands to modulate challenging drug targets that lack readily accessible and "ligandable" pockets. However, the limited membrane permeance of biologics severely restricts their intracellular applications. Moreover, different cell types may exhibit varying levels of impermeability, and some delivery vehicles might be more sensitive to this variance. Erythroid lineage cells are especially challenging to deliver cargo to because of their unique cytoskeleton and the absence of endocytosis in mature erythrocytes. We recently employed a cell permeant miniature protein to deliver bioPROTACs to human umbilical cord blood derived erythroid progenitor cells (HUDEP-2) and primary hematopoietic stem (CD34+) cells (Shen et al., ACS Cent. Sci.2022, 8, 1695-1703). While successful, the low efficiency of delivery and lack of cell-type specificity limit use of bioPROTACs in vivo. In this work, we thoroughly evaluated the performance of various recently reported cell penetrating peptides (CPPs), CPP additives, bacterial toxins, and contractile injection systems for their ability to deliver cargo to erythroid precursor cells. We also explored how targeting receptors enriched on the erythroid cell surface might improve the efficiencies and specificities of these delivery vehicles. Our results reveal that certain vehicles exhibit improved efficiencies when directed to cell surface receptors while others do not benefit from this targeting strategy. Together, these findings advance our understanding of protein delivery to challenging cell types and illustrate some of the intricacies of cell-surface receptor targeting.
© 2025 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
