Checkpoint and recombination pathways independently suppress rates of spontaneous homology-directed chromosomal translocations in budding yeast
- PMID: 40255487
- PMCID: PMC12006765
- DOI: 10.3389/fgene.2025.1479307
Checkpoint and recombination pathways independently suppress rates of spontaneous homology-directed chromosomal translocations in budding yeast
Abstract
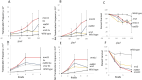
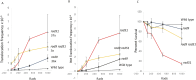
Homologous recombination between short repeated sequences, such as Alu sequences, can generate pathogenic chromosomal rearrangements. We used budding yeast to measure homologous recombination between short repeated his3 sequences located on non-homologous chromosomes to identify pathways that suppress spontaneous and radiation-associated translocations. Previous published data demonstrated that genes that participate in RAD9-mediated G2 arrest, the S phase checkpoint, and recombinational repair of double-strand breaks (DSBs) suppressed ectopic recombination between small repeats. We determined whether these pathways are independent in suppressing recombination by measuring frequencies of spontaneous recombination in single and double mutants. In the wild-type diploid, the rate of spontaneous recombination was (3 ± 1.2) × 10-8. This rate was increased 10-30-fold in the rad51, rad55, rad57, mre11, rad50, and xrs2 mutants, seven-fold in the rad9 checkpoint mutant, and 23-fold in the mec1-21 S phase checkpoint mutant. Double mutants defective in both RAD9 and in either RAD51, RAD55, or RAD57 increased spontaneous recombination rates by ∼40 fold, while double mutants defective in both the MEC1 (ATR/ATM ortholog) and RAD51 genes increased rates ∼100 fold. Compared to frequencies of radiation-associated translocations in wild type, radiation-associated frequencies increased in mre11, rad50, xrs2, rad51, rad55 and rad9 rad51 diploid mutants; an increase in radiation-associated frequencies was detected in the rad9 rad51 diploid after exposure to 100 rads X rays. These data indicate that the S phase and G2 checkpoint pathways are independent from the recombinational repair pathway in suppressing homology-directed translocations in yeast.
Keywords: budding yeast; cell cycle checkpoints; chromosomal translocations; homologous recombination; radiation.
Copyright © 2025 Zeng, Sun and Fasullo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., et al. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132. 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

