In-silico based discovery of potential Keap1 inhibitors using the strategies of pharmacophore screening, molecular docking, and MD simulation studies
- PMID: 40256239
- PMCID: PMC12008509
- DOI: 10.34172/bi.30335
In-silico based discovery of potential Keap1 inhibitors using the strategies of pharmacophore screening, molecular docking, and MD simulation studies
Abstract
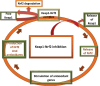
Introduction: The main objective of this research is to identify potential leads for developing potent Keap1 inhibitors.
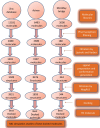
Methods: In the current research article, in-silico methods have been employed to discover potential Keap1 inhibitors. 3D-QSAR was generated using the ChemBL database of Keap1 inhibitors with IC50. The best pharmacophore was selected for the screening of three different libraries namely Asinex, MiniMaybridge, and Zinc. The molecules screened from the databases were filtered through druggability rules and molecular docking studies. The best binding molecules obtained after docking studies were subjected to physicochemical properties toxicity determination by in-silico methods. The best hits were studied for stability in the cavity of Keap1 by molecular dynamic simulations.
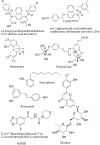
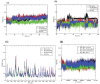
Results: The virtual screening of different databases was carried out separately and three leads, were obtained. These lead molecules ASINEX 508, MiniMaybridgeHTS_01719, and ZINC 0000952883 showed the best binding in the Keap1 cavity. The molecular dynamic simulations of the binding complexes of the leads support the docking analysis. The leads (ASINEX 508, MiniMaybridgeHTS_01719, and ZINC 0000952883) were stabilized in the Keap1 binding cavity throughout 100 ns simulation, with average RMSD values of 0.100, 0.114, and 0.106 nm, respectively.
Conclusion: This research proposes three lead molecules as potential Keap1 inhibitors based on high throughput screening, docking, and MD simulation studies. These hit molecules can be used for further design and development of Keap1 inhibitors. This research provides the preliminary data for discovering novel Keap1 inhibitors. It opens new avenues for medicinal chemists to explore antioxidant-stimulating molecules targeting the Keap1-Nrf2 pathway.
Keywords: 3DQSAR; Docking; Keap1 inhibitors; Molecular dynamics; Oxidative stress; Virtual screening.
© 2025 The Author(s).
Conflict of interest statement
Authors declare no conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Miscellaneous