Deciphering aging-associated prognosis and heterogeneity in gastric cancer through a machine learning-driven approach
- PMID: 40256325
- PMCID: PMC12008712
- DOI: 10.1016/j.isci.2025.112316
Deciphering aging-associated prognosis and heterogeneity in gastric cancer through a machine learning-driven approach
Abstract
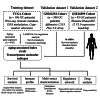
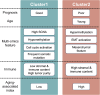
Gastric cancer (GC) is a prevalent malignancy with a high mortality rate and limited treatment options. Aging significantly contributes to tumor progression, and GC was confirmed as an aging-related heterogeneous disease. This study established an aging-associated index (AAI) using a machine learning-derived gene panel to stratify GC patients. High AAI scores associated with poor prognosis and indicated potential benefits from adjuvant chemotherapy, while showing resistance to immunotherapy. Single-cell transcriptome analysis revealed that AAI was enriched in monocyte cells within the tumor microenvironment. Two distinct molecular subtypes of GC were identified through unsupervised clustering, leading to the development of a subtype-specific regulatory network highlighting SOX7 and ELK3 as potential therapeutic targets. Drug sensitivity analyses indicated that patients with high ELK3 expression may respond to FDA-approved drugs (axitinib, dacarbazine, crizotinib, and vincristine). Finally, a user-friendly Shiny application was created to facilitate access to the prognostic model and molecular subtype classifier for GC.
Keywords: Chemistry; Computer science; Drugs.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. - PubMed
-
- Smyth E.C., Nilsson M., Grabsch H.I., van Grieken N.C., Lordick F. Gastric cancer. Lancet. 2020;396:635–648. - PubMed
-
- López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. Hallmarks of aging: An expanding universe. Cell. 2023;186:243–278. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

