Development and validation of a semi-automatic radiomics ensemble model for preoperative evaluation of breast masses in mammotome-assisted minimally invasive resection
- PMID: 40256481
- PMCID: PMC12004299
- DOI: 10.21037/gs-24-440
Development and validation of a semi-automatic radiomics ensemble model for preoperative evaluation of breast masses in mammotome-assisted minimally invasive resection
Abstract
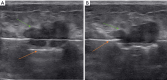
Background: Accurate preoperative differentiation of breast masses is critical for guiding individualized treatment strategies in Mammotome-assisted minimally invasive resection. While radiomics shows promise, existing methods rely on manual delineation, which is time-consuming and subjective. This study developed an ultrasound-based semi-automatic segmentation ensemble model to improve preoperative assessment.
Methods: We retrospectively analyzed preoperative ultrasound images from 773 patients (543 tumors, 230 non-tumors). Semi-automatic segmentation was performed using DeepLabv3_ResNet50 and fully convolutional network (FCN)_ResNet50. Radiomic and deep transfer learning (DTL) features were extracted to construct radiomic, deep learning, and combined models. An ensemble strategy integrated these with clinical models. Performance was evaluated via receiver operating characteristic (ROC) curves and decision curve analysis (DCA).
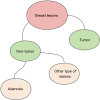
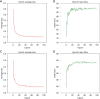
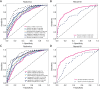
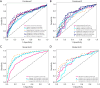
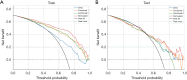
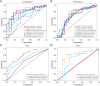
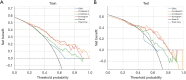
Results: The cohort included 543 tumor patients and 230 non-tumor patients (95 adenosis, 135 other benign lesions). The semi-automatic segmentation model, DeepLabv3_ResNet50, achieved a peak global accuracy of 99.4% and an average Dice coefficient of 92.0% at its best epoch. On the other hand, the FCN_ResNet50 model exhibited a peak global accuracy of 99.5% and an average Dice coefficient of 93.7% at its best epoch. In the task of predicting tumor and non-tumor patients, age, maximum diameter, and BI-RADS (Breast Imaging Reporting and Data System) classification were ultimately identified as key indicators, and the stacking model ultimately demonstrated an area under the curve (AUC) of 0.890 in the training cohort (with a sensitivity of 0.844 and a specificity of 0.815) and an AUC of 0.780 in the testing cohort (with a sensitivity of 0.713 and a specificity of 0.739). In the task of predicting adenosis and other lesion types, focus emerged as a crucial factor, and the stacking model achieved an AUC of 0.813 in the training cohort (with a sensitivity of 0.613 and a specificity of 0.859) and an AUC of 0.771 in the testing cohort (with a sensitivity of 0.759 and a specificity of 0.765).
Conclusions: Our study has established an ensemble learning model grounded in semi-automatic segmentation techniques. This model accurately distinguishes between tumor and non-tumor patients preoperatively, as well as discriminating adenosis from other lesion types among the non-tumor cohort, thus providing valuable insights for individualized treatment planning. The proposed stacking model demonstrates significant clinical utility by reducing unnecessary biopsies and saving diagnostic time compared to manual review. These improvements directly address the challenges of overtreatment and diagnostic delays in breast lesion management. By enhancing preoperative accuracy, our model supports tailored surgical planning and alleviates patient anxiety associated with indeterminate diagnoses.
Keywords: Breast lesion; deep learning; mammotome; radiomics; ultrasound (US).
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-24-440/coif). The authors have no conflicts of interest to declare.
Figures

References
LinkOut - more resources
Full Text Sources
