Rapid molecular detection of Senecavirus A based on reverse transcription loop-mediated isothermal amplification (RT-LAMP) and CRISPR/Cas12a
- PMID: 40256781
- PMCID: PMC12006090
- DOI: 10.3389/fbioe.2025.1451125
Rapid molecular detection of Senecavirus A based on reverse transcription loop-mediated isothermal amplification (RT-LAMP) and CRISPR/Cas12a
Abstract
Introduction: Senecavirus A (SVA), an emerging vesicular pathogen, is responsible for porcine idiopathic vesicular disease (PIVD). This disease is closely associated with porcine vesicular disease and acute neonatal piglet mortality, presenting a substantial threat to the global swine industry. At present, the absence of effective drugs or vaccines for treating the disease makes accurate diagnosis of SVA of paramount importance for the effective prevention and control of the disease.
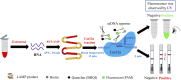
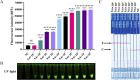
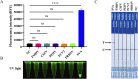
Methods: In this study, we combined reverse transcription loop-mediated isothermal amplification (RT-LAMP) and Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated protein12a (CRISPR/Cas12a) using a dual-labelled fluorescence quencher or fluorescent biotin single-stranded DNA reporter molecule to develop two rapid, reliable, and portable visual SVA assays: RT-LAMP-Cas12a-FQ and RT-LAMP-Cas12a-FB.
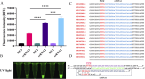
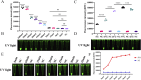
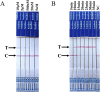
Results: The two methods exhibited comparable detection limits, with 9.6 copies/μL achieved in 40 and 45 minutes, respectively. They did not cross-react with non-target nucleic acids extracted from other related viruses and showed high specificity for SVA RNA detection. Furthermore, the methods demonstrated satisfactory performance in detecting 69 porcine adventitious samples, with no significant differences from that of quantitative reverse transcription polymerase chain reaction (RT-qPCR).
Discussion: In summary, the RT-LAMP-Cas12a-FQ and RT-LAMP-Cas12a-FB methods developed are promising for early detection and routine surveillance of porcine SVA in resource-poor areas.
Keywords: CRISPR/Cas12a; RT-LAMP; Senecavirus A; nucleic acid detection; visualisation.
Copyright © 2025 Jiang, Wang, Guo, Yang, Li, Liu, Wang, Yang, Chang and Zeng.
Conflict of interest statement
Authors CJ, RY, XL, PL, JW, JY, and YC were employed by China Agricultural Veterinary Biological Science and Technology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Specific Detection of RHDV GI.1 and GI.2 by RT-LAMP-CRISPR/Cas12a Platform.Transbound Emerg Dis. 2024 Nov 19;2024:3881457. doi: 10.1155/tbed/3881457. eCollection 2024. Transbound Emerg Dis. 2024. PMID: 40303064 Free PMC article.
-
Advancing forensic body fluid identification: A comparative analysis of RT-LAMP+CRISPR-Cas12a and established mRNA-based methods.Forensic Sci Int Genet. 2025 Jun;78:103297. doi: 10.1016/j.fsigen.2025.103297. Epub 2025 May 6. Forensic Sci Int Genet. 2025. PMID: 40347696
-
Sensitive and Visual Detection of Brassica Yellows Virus Using Reverse Transcription Loop-Mediated Isothermal Amplification-Coupled CRISPR-Cas12 Assay.Phytopathology. 2024 Feb;114(2):474-483. doi: 10.1094/PHYTO-06-23-0195-R. Epub 2024 Feb 19. Phytopathology. 2024. PMID: 37589413
-
Detection of Porcine Circovirus (PCV) Using CRISPR-Cas12a/13a Coupled with Isothermal Amplification.Viruses. 2024 Sep 30;16(10):1548. doi: 10.3390/v16101548. Viruses. 2024. PMID: 39459882 Free PMC article. Review.
-
Advances in the differential molecular diagnosis of vesicular disease pathogens in swine.Front Microbiol. 2022 Oct 25;13:1019876. doi: 10.3389/fmicb.2022.1019876. eCollection 2022. Front Microbiol. 2022. PMID: 36386633 Free PMC article. Review.
References
-
- Armson B., Walsh C., Morant N., Fowler V. L., Knowles N. J., Clark D. (2019). The development of two field‐ready reverse transcription loop-mediated isothermal amplification assays for the rapid detection of Seneca Valley virus 1. Transbound. Emerg. Dis. 66, 497–504. 10.1111/tbed.13051 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

