Single-cell landscape of peripheral immune cells in MASLD/MASH
- PMID: 40257301
- PMCID: PMC12014121
- DOI: 10.1097/HC9.0000000000000643
Single-cell landscape of peripheral immune cells in MASLD/MASH
Abstract
Background: Metabolic dysfunction-associated steatotic liver disease (MASLD) progresses to metabolic dysfunction-associated steatohepatitis (MASH) and is a major cause of liver cirrhosis. Although liver inflammation is the hallmark feature of MASH versus MASLD, the involvement of the peripheral immune cell compartments in disease progression is poorly understood, and single-cell profiles of peripheral immune cells in MASLD/MASH are not known.
Methods: Patients with MASLD/MASH and healthy volunteers have been prospectively enrolled in a cross-sectional study. Patients have been histologically stratified and further characterized by liver bulk RNA sequencing (RNA-Seq). Peripheral immune cells from patients and control blood samples have been comprehensively profiled using bulk and single RNA-Seq.
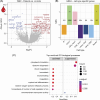
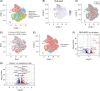
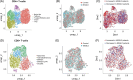
Results: Twenty-two patients with fibrosis stage less than F3 have been histologically stratified into patients with low, medium, and high disease activity scores (NAFLD activity score [NAS]). In contrast to fibrosis, the NAS group correlated with noninvasive imaging readouts and blood biomarkers of liver damage and inflammation (ALT, AST). The prevalence of type 2 diabetes and obesity increased with the NAS stage. Bulk RNA-seq profiling of patient liver biopsies revealed gene signatures that were positively and negatively associated with NAS. Known marker genes for liver fibrosis where upregulated on RNA level. Blood bulk RNA-seq showed only moderate differences in patients versus healthy controls. In contrast, single-cell analysis of white blood cells revealed multiple alterations of immune (sub-)populations, including an increased abundance of immature B cells and myeloid suppressor cells in patients with MASLD/MASH as compared to healthy controls.
Conclusions: The study gives new insights into the pathophysiology of MASLD/MASH already manifesting relatively early in peripheral immune cell compartments. This opens new avenues for the development of new biomarker diagnostics and disease therapies.
Keywords: B cells; MASH; MASLD; blood; immature immune cells; immune cells; liver; myeloid-derived suppressor cells; neutrophils; scRNA-Seq.
Copyright © 2025 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Association for the Study of Liver Diseases.
Conflict of interest statement
Agnes Anna Steixner-Kumar, Alec Dick, Dennis Böttner, Diana Santacruz, Ehsan Bahrami, Eric Simon, George Okafo, Heike Neubauer, Markus Werner, Oliver Krenkel, Tobias Geiger, Werner Rust are employees of Boehringer Ingelheim. Boehringer Ingelheim is a research-oriented pharmaceutical company. The remaining authors have no conflicts of interest.
Figures





References
-
- Younossi ZM, Blissett D, Blissett R, Henry L, Stepanova M, Younossi Y, et al. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology. 2016;64:1577–1586. - PubMed
-
- Harrison SA, Taub R. A phase 3 trial of resmetirom in NASH with liver fibrosis. N Engl J Med. 2024;390:1632–1633. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

