MyD88-mediated signaling in intestinal fibroblasts regulates macrophage antimicrobial defense and prevents dysbiosis in the gut
- PMID: 40257864
- PMCID: PMC12262590
- DOI: 10.1016/j.celrep.2025.115553
MyD88-mediated signaling in intestinal fibroblasts regulates macrophage antimicrobial defense and prevents dysbiosis in the gut
Abstract
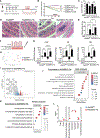
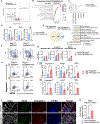
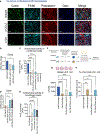
Fibroblasts that reside in the gut mucosa are among the key regulators of innate immune cells, but their role in the regulation of the defense functions of macrophages remains unknown. MyD88 is suggested to shape fibroblast responses in the intestinal microenvironment. We found that mice lacking MyD88 in fibroblasts showed a decrease in the colonic antimicrobial defense, developing dysbiosis and aggravated dextran sulfate sodium (DSS)-induced colitis. These pathological changes were associated with the accumulation of Arginase 1+ macrophages with low antimicrobial defense capability. Mechanistically, the production of interleukin (IL)-6 and CCL2 downstream of MyD88 was critically involved in fibroblast-mediated support of macrophage antimicrobial function, and IL-6/CCL2 neutralization resulted in the generation of macrophages with decreased production of the antimicrobial peptide cathelicidin and impaired bacterial clearance. Collectively, these findings revealed a critical role of fibroblast-intrinsic MyD88 signaling in regulating macrophage antimicrobial defense under colonic homeostasis, and its disruption results in dysbiosis, predisposing the host to the development of intestinal inflammation.
Keywords: CP: Immunology; MyD88; antimicrobial defense; dysbiosis; fibroblasts; gut; macrophages.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Similar articles
-
Gut microbial Nordihydroguaiaretic acid suppresses macrophage pyroptosis to regulate epithelial homeostasis and inflammation.Gut Microbes. 2025 Dec;17(1):2518338. doi: 10.1080/19490976.2025.2518338. Epub 2025 Jul 1. Gut Microbes. 2025. PMID: 40596758 Free PMC article.
-
Gut microbiome promotes succinate-induced ulcerative colitis by enhancing glycolysis through SUCNR1/NF-κB signaling pathway.Am J Physiol Cell Physiol. 2025 Aug 1;329(2):C440-C454. doi: 10.1152/ajpcell.00411.2025. Epub 2025 Jun 23. Am J Physiol Cell Physiol. 2025. PMID: 40549551
-
The effects of candidate probiotic strains on the gut environment in dextran sulfate sodium-induced colitis mouse.Sci Rep. 2025 Jul 1;15(1):21103. doi: 10.1038/s41598-025-03860-5. Sci Rep. 2025. PMID: 40593990 Free PMC article.
-
Role of the intestinal flora-immunity axis in the pathogenesis of rheumatoid arthritis-mechanisms regulating short-chain fatty acids and Th17/Treg homeostasis.Mol Biol Rep. 2025 Jun 21;52(1):617. doi: 10.1007/s11033-025-10714-w. Mol Biol Rep. 2025. PMID: 40544212 Review.
-
Synbiotics, prebiotics and probiotics for solid organ transplant recipients.Cochrane Database Syst Rev. 2022 Sep 20;9(9):CD014804. doi: 10.1002/14651858.CD014804.pub2. Cochrane Database Syst Rev. 2022. PMID: 36126902 Free PMC article.
References
-
- Alatab S, Sepanlou SG, Ikuta K, Vahedi H, Bisignano C, Safiri S, Sadeghi A, Nixon MR, Abdoli A, Abolhassani H, and Alipour V (2020). The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet Gastroenterology & Hepatology 5, 17–30. - PMC - PubMed
-
- Pizarro TT, Stappenbeck TS, Rieder F, Rosen MJ, Colombel JF, Donowitz M, Towne J, Mazmanian SK, Faith JJ, Hodin RA, et al. (2019). Challenges in IBD Research: Preclinical Human IBD Mechanisms. Inflamm. Bowel Dis 25, S5–S12. - PubMed
-
- Uhlig HH, and Powrie F (2018). Translating Immunology into Therapeutic Concepts for Inflammatory Bowel Disease. Annu. Rev. Immunol 36, 755–781. - PubMed
-
- Ignacio A, Czyz S, and McCoy KD (2024). Early life microbiome influences on development of the mucosal innate immune system. Semin. Immunol 73, 101885. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

