Addressing data management and analysis challenges in viral genomics: The Swiss HIV cohort study viral next generation sequencing database
- PMID: 40257980
- PMCID: PMC12011223
- DOI: 10.1371/journal.pdig.0000825
Addressing data management and analysis challenges in viral genomics: The Swiss HIV cohort study viral next generation sequencing database
Abstract
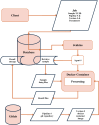
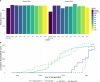
Numerous HIV related outcomes can be determined on the viral genome, for example, resistance associated mutations, population transmission dynamics, viral heritability traits, or time since infection. Viral sequences of people with HIV (PWH) are therefore essential for therapeutic and research purposes. While in the first three decades of the HIV pandemic viral genomes were mainly sequenced using Sanger sequencing, the last decade has seen a shift towards next-generation sequencing (NGS) as the preferred method. NGS can achieve near full length genome sequence coverage and simultaneously, it accurately encapsulates the within-host diversity by characterizing HIV subpopulations. NGS opens new avenues for HIV research, but it also presents challenges concerning data management and analysis. We therefore set up the Swiss HIV Cohort Study Viral NGS Database (SHCND) to address key issues in the handling of NGS data including high loads of raw- and processed NGS data, data storage solutions, downstream application of sophisticated bioinformatic tools, high-performance computing resources, and reproducibility. The database is nested within the Swiss HIV Cohort Study (SHCS) and the Zurich Primary HIV Infection Cohort Study (ZPHI), which together enrolled 21,876 PWH since 1988 and include a biobank dating back to the early nineties. Since its initiation in 2018, the SHCND accumulated NGS sequences (plasma and proviral origin) of 5,178 unique PWH. We here describe the design, set-up, and use of this NGS database. Overall, the SHCND has contributed to several research projects on HIV pathogenesis, treatment, drug resistance, and molecular epidemiology, and has thereby become a central part of HIV-genomics research in Switzerland.
Copyright: © 2025 Zeeb et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: Within the last 5 years, K.J.M. has received travel grants and advisory board honoraria from Gilead Sciences and ViiV; and the University of Zurich received research grants from Gilead Sciences and Novartis for studies that Dr Metzner serves as principal investigator. H. F. G. has received research grants from the Swiss National Science Foundation, Swiss HIV Cohort Study, Yvonne Jacob Foundation, NIH, Gilead, ViiV, and is a subcontractor to a Bill and Melinda Gates foundation grant, paid to his institution; personal honoraria for data safety monitoring board or advisory board consultations from Merck, ViiV healthcare, Gilead Sciences, Janssen, Johnson and Johnson, Novartis, and GSK; and personal travel expenses from Gilead. All other authors declare no conflicts of interest.
Figures


Similar articles
-
Frequency matters: comparison of drug resistance mutation detection by Sanger and next-generation sequencing in HIV-1.J Antimicrob Chemother. 2023 Mar 2;78(3):656-664. doi: 10.1093/jac/dkac430. J Antimicrob Chemother. 2023. PMID: 36738248
-
Bioinformatic data processing pipelines in support of next-generation sequencing-based HIV drug resistance testing: the Winnipeg Consensus.J Int AIDS Soc. 2018 Oct;21(10):e25193. doi: 10.1002/jia2.25193. J Int AIDS Soc. 2018. PMID: 30350345 Free PMC article.
-
New Aspects of the Virus Life Cycle and Clinical Utility of Next Generation Sequencing based HIV-1 Resistance Testing in the Genomic, the Proviral, and the Viral Reservoir of Peripheral Blood Mononuclear Cells.Curr HIV Res. 2022;20(3):213-221. doi: 10.2174/1570162X20666220324111418. Curr HIV Res. 2022. PMID: 35331114
-
Technologies for HIV-1 drug resistance testing: inventory and needs.Curr Opin HIV AIDS. 2022 Jul 1;17(4):222-228. doi: 10.1097/COH.0000000000000737. Curr Opin HIV AIDS. 2022. PMID: 35762377 Review.
-
Probe Capture Enrichment Methods for HIV and HCV Genome Sequencing and Drug Resistance Genotyping.Pathogens. 2022 Jun 16;11(6):693. doi: 10.3390/pathogens11060693. Pathogens. 2022. PMID: 35745547 Free PMC article. Review.
Cited by
-
Phylogenetics and molecular evolution to understand and curb the HIV pandemic.Nat Rev Microbiol. 2025 Jun 30. doi: 10.1038/s41579-025-01202-w. Online ahead of print. Nat Rev Microbiol. 2025. PMID: 40588586 Review.
References
-
- Chabria S, Gupta S, Kozal M. Deep sequencing of HIV: clinical and research applications. Annual Review of Genomics and Human Genetics. 2014;15:295–325. https://pubmed.ncbi.nlm.nih.gov/24821496/ - PubMed
-
- Bonsall D, Golubchik T, de Cesare M, Limbada M, Kosloff B, MacIntyre-Cockett G, et al.. A comprehensive genomics solution for HIV surveillance and clinical monitoring in low-income settings. Journal of Clinical Microbiology. 2020;58(10):382–402. https://pmc/articles/PMC7512176/ - PMC - PubMed
LinkOut - more resources
Full Text Sources
