Isolating selective from non-selective forces using site frequency ratios
- PMID: 40258089
- PMCID: PMC12064048
- DOI: 10.1371/journal.pgen.1011427
Isolating selective from non-selective forces using site frequency ratios
Abstract
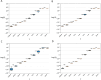
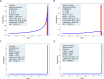
A new method is introduced for estimating the distribution of mutation fitness effects using site frequency spectra. Unlike previous methods, which make assumptions about non-selective factors, or that try to incorporate such factors into the underlying model, this new method mostly avoids non-selective effects by working with the ratios of counts of selected sites to neutral sites. An expression for the likelihood of a set of selected/neutral ratios is found by treating the ratio of two Poisson random variables as the ratio of two gaussian random variables. This approach also avoids the need to estimate the relative mutation rates of selected and neutral sites. Simulations over a wide range of demographic models, with linked selection effects show that the new SFRatios method performs well for statistical tests of selection, and it performs well for estimating the distribution of selection effects. Performance was better with weak selection models and for expansion and structured demographic models than for bottleneck models. Applications to two populations of Drosophila melanogaster reveal clear but very weak selection on synonymous sites. For nonsynonymous sites, selection was found to be consistent with previous estimates and stronger for an African population than for one from North Carolina.
Copyright: © 2025 Hey. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

