Rab32 regulates Golgi structure and cell migration through Protein Kinase A-mediated phosphorylation of Optineurin
- PMID: 40258145
- PMCID: PMC12054839
- DOI: 10.1073/pnas.2502971122
Rab32 regulates Golgi structure and cell migration through Protein Kinase A-mediated phosphorylation of Optineurin
Abstract
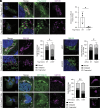
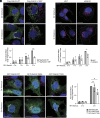
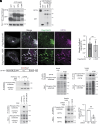
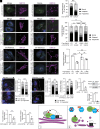
Rab32 is a small GTPase and molecular switch implicated in vesicular trafficking. Rab32 is also an A-Kinase Anchoring Protein (AKAP), which anchors cAMP-dependent Protein Kinase (PKA) to specific subcellular locations and specifies PKA phosphorylation of nearby substrates. Surprisingly, we found that a form of Rab32 deficient in PKA binding (Rab32 L188P) relocalized away from the Golgi apparatus and induced a marked disruption in Golgi organization, assembly, and dynamics. Although Rab32 L188P did not cause a global defect in PKA activity, our data indicate that Rab32 facilitates the phosphorylation of a specific PKA substrate. We uncovered a direct interaction between Rab32 and the adaptor protein optineurin (OPTN), which regulates Golgi dynamics. Further, our data indicate that optineurin is phosphorylated by PKA at Ser342 in a Rab32-dependent manner. Critically, blocking phosphorylation at OPTN Ser342 leads to Golgi fragmentation, and a phospho-mimetic version of OPTN rescues Golgi defects induced by Rab32 L188P. Finally, Rab32 AKAP function and OPTN phosphorylation are required for Golgi repositioning during cell migration, contributing to tumor cell invasion. Together, these data reveal a role for Rab32 in regulating Golgi dynamics through PKA-mediated phosphorylation of OPTN.
Keywords: Golgi; Optineurin; Protein Kinase A; Rab32; migration.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
References
-
- Stenmark H., Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 (2009). - PubMed
MeSH terms
Substances
Grants and funding
- S10 OD028633/OD/NIH HHS/United States
- P50CA210964/HHS | NIH | National Cancer Institute (NCI)
- P30CA015083/HHS | NIH | National Cancer Institute (NCI)
- S10OD028633/HHS | NIH | NIH Office of the Director (OD)
- P30DK084567/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
LinkOut - more resources
Full Text Sources

