Chromosome-level genome assembly and annotation of Chinese herring (Ilisha elongata)
- PMID: 40258812
- PMCID: PMC12012206
- DOI: 10.1038/s41597-025-04790-7
Chromosome-level genome assembly and annotation of Chinese herring (Ilisha elongata)
Abstract
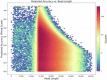
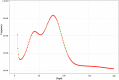
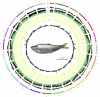
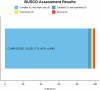
The Chinese herring (Ilisha elongata) is a commercially and scientifically significant fish species. In this study, we conducted high-precision whole-genome sequencing using two high-throughput platforms: second-generation MGI and third-generation PacBio. Hi-C technology assisted in assembling the contig sequences onto 24 chromosomes, resulting in a high-quality chromosome-level genome map with excellent continuity and coverage. The completed genome size was approximately 815 Mb, with a contig N50 of 4.82 Mb, scaffold N50 of 32.61 Mb, and a chromosome mounting rate of 95.32%. SNP and InDel purity rates were 0.003% and 0.012%, respectively, and the genome assembly completeness was 96.68%, assessed by BUSCO. Repetitive sequences were annotated via ab initio and homology predictions, identifying 295.7 Mb of repetitive sequences, constituting 35.08% of the genome. A total of 26,381 protein-coding genes were predicted, with 24,596 functionally annotated.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Genome sequence of the barred knifejaw Oplegnathus fasciatus (Temminck & Schlegel, 1844): the first chromosome-level draft genome in the family Oplegnathidae.Gigascience. 2019 Mar 1;8(3):giz013. doi: 10.1093/gigascience/giz013. Gigascience. 2019. PMID: 30715332 Free PMC article.
-
A chromosome-level genome assembly of the Chinese herring (Ilisha elongata) uncovered its population dynamics and genes related to lipid metabolism.J Hered. 2025 May 14:esaf028. doi: 10.1093/jhered/esaf028. Online ahead of print. J Hered. 2025. PMID: 40360201
-
A Chromosome-Level Genome Assembly of the Dark Sleeper Odontobutis potamophila.Genome Biol Evol. 2021 Feb 3;13(2):evaa271. doi: 10.1093/gbe/evaa271. Genome Biol Evol. 2021. PMID: 33576781 Free PMC article.
-
Chromosome-level genome assembly of Acrossocheilus fasciatus using PacBio sequencing and Hi-C technology.Sci Data. 2024 Feb 3;11(1):166. doi: 10.1038/s41597-024-02999-6. Sci Data. 2024. PMID: 38310107 Free PMC article.
-
An improved chromosome-level genome assembly and annotation of Echeneis naucrates.Sci Data. 2024 May 4;11(1):452. doi: 10.1038/s41597-024-03309-w. Sci Data. 2024. PMID: 38704456 Free PMC article.
References
-
- Zhang, J., Takita, T. & Zhang, C. Reproductive biology of Ilisha elongata (Teleostei: Pristigasteridae) in Ariake Sound, Japan: Implications for estuarine fish conservation in Asia. Estuarine, Coastal and Shelf Science81, 105–113, 10.1016/j.ecss.2008.10.013 (2009).
-
- Whitehead, P. J. Clupeoid fishes of the World (Suborder Clupeoidei): An annotated and illustrated catalogue of the herrings, sardines, pilchards, sprats, shads, anchovies and wolf herrings. Part 1. Chirocentridae, Clupeidae and Pristigasteridae. FAO Fisheries Synopsis125, 1, 10.1093/ices/fct001 (1985).
-
- Wang, X. et al. Early life history of Ilisha elongata (Pristigasteridae, Clupeiformes, Pisces) in Ariake Sound, Shimabara Bay, Japan. Plankton and Benthos Research16, 210–220, 10.3800/pbr.16.210 (2021).
-
- Cossins, A. R. & Crawford, D. L. Fish as models for environmental genomics. Nature Reviews Genetics6, 324–333 (2005). - PubMed
-
- Kasahara, M. et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 447, 714-719 (2007). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

