Integrated multiomics analysis identifies potential biomarkers and therapeutic targets for autophagy associated AKI to CKD transition
- PMID: 40258914
- PMCID: PMC12012120
- DOI: 10.1038/s41598-025-97269-9
Integrated multiomics analysis identifies potential biomarkers and therapeutic targets for autophagy associated AKI to CKD transition
Abstract
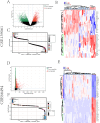
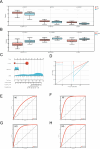
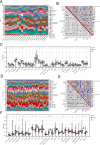
This study explored the relationship between acute kidney injury (AKI) and chronic kidney disease (CKD), focusing on autophagy-related genes and their immune infiltration during the transition from AKI to CKD. We performed weighted correlation network analysis (WGCNA) using two microarray datasets (GSE139061 and GSE66494) in the GEO database and identified autophagy signatures by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG), and GSEA enrichment analysis. Machine learning algorithms such as LASSO, random forest, and XGBoost were used to construct the diagnostic model, and the diagnostic performance of GSE30718 (AKI) and GSE37171 (CKD) was used as validation cohorts to evaluate its diagnostic performance. The study identified 14 autophagy candidate genes, among which ATP6V1C1 and COPA were identified as key biomarkers that were able to effectively distinguish between AKI and CKD. Immune cell infiltration and GSEA analysis revealed immune dysregulation in AKI, and these genes were associated with inflammation and immune pathways. Single-cell analysis showed that ATP6V1C1 and COPA were specifically expressed in AKI and CKD, which may be related to renal fibrosis. In addition, drug prediction and molecular docking analysis proposed SZ(+)-(S)-202-791 and PDE4 inhibitor 16 as potential therapeutic agents. In summary, this study provides new insights into the relationship between AKI and CKD and lays a foundation for the development of new treatment strategies.
Keywords: AKI; CKD; Autophagy; Biomarkers; Machine learning.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics statement: This study does not involve human and animal experiments and does not require ethical approval.
Figures






Similar articles
-
Identification of common molecular signatures of SARS-CoV-2 infection and its influence on acute kidney injury and chronic kidney disease.Front Immunol. 2023 Mar 21;14:961642. doi: 10.3389/fimmu.2023.961642. eCollection 2023. Front Immunol. 2023. PMID: 37026010 Free PMC article.
-
Augmenter of liver regeneration inhibits renal fibrosis during acute kidney injury to chronic kidney disease transition by regulating autophagic flux.Arch Biochem Biophys. 2025 Feb;764:110218. doi: 10.1016/j.abb.2024.110218. Epub 2024 Nov 27. Arch Biochem Biophys. 2025. PMID: 39613286
-
Identification of Important Modules and Hub Gene in Chronic Kidney Disease Based on WGCNA.J Immunol Res. 2022 May 4;2022:4615292. doi: 10.1155/2022/4615292. eCollection 2022. J Immunol Res. 2022. PMID: 35571562 Free PMC article.
-
Role of endoplasmic reticulum stress and autophagy in the transition from acute kidney injury to chronic kidney disease.J Cell Physiol. 2023 Jan;238(1):82-93. doi: 10.1002/jcp.30918. Epub 2022 Nov 21. J Cell Physiol. 2023. PMID: 36409755 Review.
-
The Role of Mitochondria in Acute Kidney Injury and Chronic Kidney Disease and Its Therapeutic Potential.Int J Mol Sci. 2021 Oct 19;22(20):11253. doi: 10.3390/ijms222011253. Int J Mol Sci. 2021. PMID: 34681922 Free PMC article. Review.
References
-
- Bucaloiu, I. D. et al. Increased risk of death and de Novo chronic kidney disease following reversible acute kidney injury[J]. Kidney Int.81 (5), 477–485 (2012). - PubMed
-
- Menon, S., Symons, J. M. & Selewski, D. T. Acute kidney Injury[J]. Pediatr. Rev.44 (5), 265–279 (2023). - PubMed
-
- See, E. J. et al. Long-term risk of adverse outcomes after acute kidney injury: a systematic review and meta-analysis of cohort studies using consensus definitions of exposure[J]. Kidney Int.95 (1), 160–172 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

