Viral metagenomics reveals diverse viruses in the fecal samples of children with acute respiratory infection
- PMID: 40260089
- PMCID: PMC12009832
- DOI: 10.3389/fmicb.2025.1564755
Viral metagenomics reveals diverse viruses in the fecal samples of children with acute respiratory infection
Abstract
Introduction: Changes in the gut microbiome have been associated with the development of acute respiratory infection (ARI). However, due to methodological limitations, our knowledge of the gut virome in patients with ARIs remains limited.
Methods: In this study, fecal samples from children with ARI were investigated using viral metagenomics.
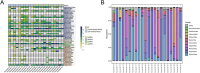
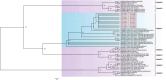
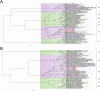
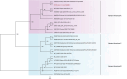
Results: The fecal virome was analyzed, and several suspected disease-causing viruses were identified. The five viral families with the highest abundance of sequence reads were Podoviridae, Virgaviridae, Siphoviridae, Microviridae, and Myoviridae. Additionally, human adenovirus, human bocavirus, human astrovirus, norovirus, and human rhinovirus were detected. The genome sequences of these viruses were respectively described, and phylogenetic trees were constructed using the gene sequences of the viruses.
Discussion: We characterized the composition of gut virome in children with acute respiratory infections. However, further research is required to elucidate the relationship between acute respiratory infection and gut viruses.
Keywords: acute respiratory infection; children; fecal samples; viral metagenomics; virus evolution.
Copyright © 2025 Xu, Pan, Yuan, Zhu, Wei, Lu and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Alves J., Teixeira D., Siqueira J., Deus D., Oliveira D., Ferreira J., et al. (2024). Epidemiology and molecular detection of human adenovirus and non-polio enterovirus in fecal samples of children with acute gastroenteritis: A five-year surveillance in northern Brazil. PLoS One 19:e0296568. 10.1371/journal.pone.0296568 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

