Glucose deprivation and identification of TXNIP as an immunometabolic modulator of T cell activation in cancer
- PMID: 40260243
- PMCID: PMC12010123
- DOI: 10.3389/fimmu.2025.1548509
Glucose deprivation and identification of TXNIP as an immunometabolic modulator of T cell activation in cancer
Abstract
Background: The ability of immune cells to rapidly respond to pathogens or malignant cells is tightly linked to metabolic pathways. In cancer, the tumor microenvironment (TME) represents a complex system with a strong metabolism stress, in part due to glucose shortage, which limits proper T cell activation, differentiation and functions preventing anti-tumor immunity.
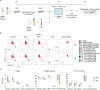
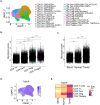
Methods: In this study, we evaluated T cell immune reactivity in glucose-restricted mixed lymphocyte reaction (MLR), using a comprehensive profiling of soluble factors, multiparametric flow cytometry and single cell RNA sequencing (scRNA-seq).
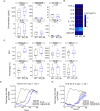
Results: We determined that glucose restriction potentiates anti-PD-1 immune responses and identified thioredoxin-interacting protein (TXNIP), a negative regulator of glucose uptake, as a potential immunometabolic modulator of T cell activation. We confirmed TXNIP downregulation in tumor infiltrating T cells in cancer patients. We next investigated the implication of TXNIP in modulating immune effector functions in primary human T cells and showed that TXNIP depletion increased IFN-γ secretion and tumor cell killing.
Conclusions: TXNIP is at the interface between immunometabolism and T cell activation and could represent a potential target for immuno-oncology treatments.
Keywords: T cells activation; TXNIP; cancer immunotherapy; glucose deprivation; mixed lymphocyte reaction; single-cell RNA-sequencing; tumor microenvironment.
Copyright © 2025 Dubuisson, Mangelinck, Knockaert, Zichi, Becht, Philippon, Dromaint-Catesson, Fasquel, Melchiore, Provost, Walas, Darville, Galizzi, Lefebvre, Blanc and Lombardi.
Conflict of interest statement
All authors were employed by company Servier. The authors declare that this study received funding from Servier. The funder had the following involvement in the study: study design, data collection and analysis, decision to publish and preparation of the manuscript.
Figures




References
-
- Bain B, Vas M, Lowenstein L. Reaction between leukocytes in mixed peripheral blood cultures. In: Federation Proceedings. FEDERATION AMER SOC EXP BIOL 9650 ROCKVILLE PIKE. BETHESDA, MD; (1963). p. 428–.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

