Staphylococcus epidermidis uses the SrrAB regulatory system to modulate oxidative stress and intracellular survival in mouse macrophage cell line Ana-1
- PMID: 40261004
- PMCID: PMC12090800
- DOI: 10.1128/msystems.01737-24
Staphylococcus epidermidis uses the SrrAB regulatory system to modulate oxidative stress and intracellular survival in mouse macrophage cell line Ana-1
Abstract
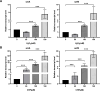
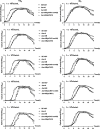
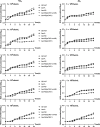
The two-component system (TCS) SrrAB responds to oxidative stress in Staphylococcus epidermidis. A srrAB deletion mutant (∆srrAB) was constructed using S. epidermidis strain 1457 (SE1457) as the parent strain to study its regulatory function in oxidative stress. Compared to SE1457, the viable cell counts of the ∆srrAB mutant significantly decreased in the post-stationary phase culture, coinciding with a sharp increase in reactive oxidative species (ROS) accumulation. The impaired growth of the ∆srrAB mutant was partially restored by shifting the culture from oxic to microaerobic conditions. Consistently, growth of the ∆srrAB mutant in tryptone soy broth (TSB) medium containing H2O2 was notably inhibited compared to parent strain SE1457, and the mutant showed significantly decreased resistance (100- to 1,000-fold) to H2O2 and cumene hydroperoxide in both oxic and microaerobic conditions, which was fully rescued by the addition of ROS inhibitor 2,2-dipyridyl. Furthermore, the deletion of srrAB resulted in decreased intracellular survival in the Ana-1 macrophages, likely due to intracellular ROS accumulation. The complementation of srrAB in the ∆srrAB mutant restored ROS resistance and intracellular survival to wild-type levels. RNA-seq analysis revealed that srrAB deletion affected the transcription levels of 610 genes, including those involved in oxidative stress, respiratory and energy metabolism, and transition ion homeostasis. These findings were corroborated by quantitative real-time reverse transcription-PCR. In the ∆srrAB mutant, expressions of ROS-scavenging genes katA, ahpC, scdA, serp1797, and serp0483 were downregulated compared to SE1457. Electrophoretic mobility shift assay further demonstrated phosphorylated SrrA bound to the promoter regions of srrAB, katA, ahpC, scdA, serp1797, and serp0483 genes. This study elucidates that in S. epidermidis, SrrAB is the critical TCS to sense and respond to the oxidants, directly regulating transcription levels of the genes involved in ROS scavenging and ion homeostasis, thereby facilitating S. epidermidis detoxification of ROS and adaptation to the commensal environment.
Importance: Staphylococcus epidermidis in the human skin and mucous microbiome is a leading cause of hospital-acquired infection, whereas the mechanism by which it inhabits, adapts, and further results in infection is not well known. In this study, we found that the two-component regulatory system SrrAB directly regulates transcription levels of the genes involved in reactive oxidative species (ROS) scavenging and ion homeostasis in S. epidermidis, influencing ROS accumulation during growth, thereby facilitating detoxification of ROS and adaptation to the commensal environment. This work provides new molecular insight into the mechanisms of SrrAB in regulating resistance and intracellular viability against oxidative stress in S. epidermidis.
Keywords: ROS; Staphylococcal respiratory response; Staphylococcus epidermidis; macrophage; oxidative stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Staphylococcus epidermidis SrrAB regulates bacterial growth and biofilm formation differently under oxic and microaerobic conditions.J Bacteriol. 2015 Feb;197(3):459-76. doi: 10.1128/JB.02231-14. Epub 2014 Nov 17. J Bacteriol. 2015. PMID: 25404696 Free PMC article.
-
The Vancomycin Resistance-Associated Regulatory System VraSR Modulates Biofilm Formation of Staphylococcus epidermidis in an ica-Dependent Manner.mSphere. 2021 Oct 27;6(5):e0064121. doi: 10.1128/mSphere.00641-21. Epub 2021 Sep 22. mSphere. 2021. PMID: 34550006 Free PMC article.
-
The Staphylococcus aureus SrrAB Regulatory System Modulates Hydrogen Peroxide Resistance Factors, Which Imparts Protection to Aconitase during Aerobic Growth.PLoS One. 2017 Jan 18;12(1):e0170283. doi: 10.1371/journal.pone.0170283. eCollection 2017. PLoS One. 2017. PMID: 28099473 Free PMC article.
-
The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia.mBio. 2013 Nov 12;4(6):e00696-13. doi: 10.1128/mBio.00696-13. mBio. 2013. PMID: 24222487 Free PMC article.
-
The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions.Mol Microbiol. 2007 Sep;65(5):1276-87. doi: 10.1111/j.1365-2958.2007.05863.x. Mol Microbiol. 2007. PMID: 17697253
References
-
- Amer MA, Darwish MM, Soliman NS, Amin HM. 2024. Resistome, mobilome, and virulome explored in clinical isolates derived from acne patients in Egypt: unveiling unique traits of an emerging coagulase-negative Staphylococcus pathogen. Front Cell Infect Microbiol 14:1328390. doi:10.3389/fcimb.2024.1328390 - DOI - PMC - PubMed
-
- Maduka-Ezeh AN, Greenwood-Quaintance KE, Karau MJ, Berbari EF, Osmon DR, Hanssen AD, Steckelberg JM, Patel R. 2012. Antimicrobial susceptibility and biofilm formation of Staphylococcus epidermidis small colony variants associated with prosthetic joint infection. Diagn Microbiol Infect Dis 74:224–229. doi:10.1016/j.diagmicrobio.2012.06.029 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
