Cholesterol 25-hydroxylase inhibits Newcastle disease virus replication by enzyme activity-dependent and direct interaction with nucleocapsid protein
- PMID: 40261014
- PMCID: PMC12090710
- DOI: 10.1128/jvi.00428-25
Cholesterol 25-hydroxylase inhibits Newcastle disease virus replication by enzyme activity-dependent and direct interaction with nucleocapsid protein
Abstract
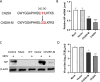
Newcastle disease virus (NDV) is a significant enveloped virus within the Paramyxoviridae family, posing a major threat to the global poultry industry. Increasing evidence suggests that cholesterol-25-hydroxylase (CH25H) and its enzymatic product, 25-hydroxycholesterol (25HC), exhibit broad-spectrum antiviral activity properties by modulating lipid metabolism and various signaling pathways. However, the specific role of CH25H in regulating NDV infection and replication remains unclear. In this study, we demonstrate that CH25H significantly inhibits NDV replication by blocking viral entry through its enzymatic product, 25HC. Notably, a catalytic mutant of CH25H (CH25H-M), which lacks hydroxylase activity, still retains partial ability to inhibit NDV replication, suggesting the involvement of an enzyme-independent antiviral mechanism. Furthermore, we found that CH25H interacts with the viral nucleoprotein (NP), leading to a reduction in NP expression and inhibition of viral ribonucleoprotein (RNP) complex activity. These findings reveal that CH25H exerts antiviral effects through both enzyme-dependent and -independent mechanisms, providing new insights into its role in host defense and offering potential targets for the development of antiviral therapies.IMPORTANCECholesterol 25-hydroxylase (CH25H) is a multifunctional host protein that has been implicated in regulating the life cycles of various viruses. As a prototype of paramyxovirus, Newcastle disease virus (NDV) poses a significant threat to the global poultry industry, causing substantial economic losses. Uncovering the mechanisms of NDV-host interactions is crucial for unraveling the viral pathogenesis and the host immune response, which can drive the development of effective antiviral therapies. Here, we demonstrate that CH25H, whose expression is induced upon NDV infection, plays a pivotal role in restricting viral replication. Specifically, CH25H interacts with the viral NP and inhibits the viral RNP activity. These findings expand our understanding of CH25H's antiviral functions and offer new insights into virus-host interactions, providing potential targets for the development of novel antiviral drugs against NDV and related pathogens.
Keywords: 25-hydroxycholesterol; Newcastle disease virus; cholesterol 25-hydroxylase; viral replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




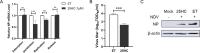
Similar articles
-
Cholesterol 25-hydroxylase inhibits Newcastle disease virus replication by its architectural damage and blocking HN protein.Antiviral Res. 2025 Sep;241:106239. doi: 10.1016/j.antiviral.2025.106239. Epub 2025 Jul 18. Antiviral Res. 2025. PMID: 40685090
-
Cholesterol 25-Hydroxylase Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication through Enzyme Activity-Dependent and -Independent Mechanisms.J Virol. 2017 Sep 12;91(19):e00827-17. doi: 10.1128/JVI.00827-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28724759 Free PMC article.
-
Cholesterol 25-hydroxylase negatively regulates porcine intestinal coronavirus replication by the production of 25-hydroxycholesterol.Vet Microbiol. 2019 Apr;231:129-138. doi: 10.1016/j.vetmic.2019.03.004. Epub 2019 Mar 6. Vet Microbiol. 2019. PMID: 30955800 Free PMC article.
-
The role of cholesterol 25-hydroxylase in viral infections: Mechanisms and implications.Pathol Res Pract. 2023 Sep;249:154783. doi: 10.1016/j.prp.2023.154783. Epub 2023 Aug 24. Pathol Res Pract. 2023. PMID: 37660656 Review.
-
Recent advancements in the diverse roles of polymerase-associated proteins in the replication and pathogenesis of Newcastle disease virus.Vet Res. 2025 Jan 12;56(1):8. doi: 10.1186/s13567-024-01429-0. Vet Res. 2025. PMID: 39800751 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- 20220101314JC/Department of Science and Technology of Jilin Province
- YDZJ202102CXJD029/Department of Science and Technology of Jilin Province
- YDZJ202301ZYTS326/Department of Science and Technology of Jilin Province
- U21A20261/National Natural Science Foundation of China
- 32473121/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

