Comparative genomic analysis of 255 Oenococcus oeni isolates from China: unveiling strain diversity and genotype-phenotype associations of acid resistance
- PMID: 40261018
- PMCID: PMC12131864
- DOI: 10.1128/spectrum.03265-24
Comparative genomic analysis of 255 Oenococcus oeni isolates from China: unveiling strain diversity and genotype-phenotype associations of acid resistance
Abstract
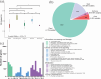
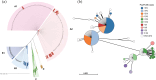
Oenococcus oeni, the only species of lactic acid bacteria capable of fully completing malolactic fermentation under challenging wine conditions, continues to intrigue researchers owing to its remarkable adaptability, particularly in combating acid stress. However, the mechanism underlying its superior adaptation to wine stresses still remains elusive due to the lack of viable genetic manipulation tools for this species. In this study, we conducted genomic sequencing and acid resistance phenotype analysis of 255 O. oeni isolates derived from diverse wine regions across China, aiming to elucidate their strain diversity and genotype-phenotype associations of acid resistance through comparative genomics. A significant correlation between phenotypes and evolutionary relationships was observed. Notably, phylogroup B predominantly consisted of acid-resistant isolates, primarily originating from Shandong and Shaanxi wine regions. Furthermore, we uncovered a noteworthy linkage between prophage genomic islands and acid resistance phenotype. Using genome-wide association studies, we identified key genes correlated with acid resistance, primarily involved in carbohydrates and amino acid metabolism processes. This study offers profound insights into the genetic diversity and genetic basis underlying adaptation mechanisms to acid stress in O. oeni.IMPORTANCEThis study provides valuable insights into the genetic basis of acid resistance in Oenococcus oeni, a key lactic acid bacterium in winemaking. By analyzing 255 isolates from diverse wine regions in China, we identified significant correlations between strain diversity, genomic islands, and acid resistance phenotypes. Our findings reveal that certain prophage-related genomic islands and specific genes are closely linked to acid resistance, offering a deeper understanding of how O. oeni adapts to acidic environments. These discoveries not only advance our knowledge of microbial stress responses but also pave the way for selecting and engineering acid-resistant strains, enhancing malolactic fermentation efficiency and wine quality. This research underscores the importance of genomics in improving winemaking practices and addressing challenges posed by high-acidity wines.
Keywords: genome-wide association study; genomic island; horizontal gene transfer; lactic acid bacteria; pan-genome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Identification of variable genomic regions related to stress response in Oenococcus oeni.Food Res Int. 2017 Dec;102:625-638. doi: 10.1016/j.foodres.2017.09.039. Epub 2017 Sep 19. Food Res Int. 2017. PMID: 29195994
-
Comparative genomics and phenotypes of Oenococcus alcoholitolerans and Oenococcus oeni, two sister species adapted to ethanol-rich environments.Food Microbiol. 2026 Jan;133:104886. doi: 10.1016/j.fm.2025.104886. Epub 2025 Jul 28. Food Microbiol. 2026. PMID: 40889861
-
Genomic diversity of Oenococcus oeni populations from Castilla La Mancha and La Rioja Tempranillo red wines.Food Microbiol. 2015 Aug;49:82-94. doi: 10.1016/j.fm.2015.02.001. Epub 2015 Feb 12. Food Microbiol. 2015. PMID: 25846918
-
Distribution of Oenococcus oeni populations in natural habitats.Appl Microbiol Biotechnol. 2019 Apr;103(7):2937-2945. doi: 10.1007/s00253-019-09689-z. Epub 2019 Feb 20. Appl Microbiol Biotechnol. 2019. PMID: 30788540 Free PMC article. Review.
-
Genomic variations of Oenococcus oeni strains and the potential to impact on malolactic fermentation and aroma compounds in wine.Appl Microbiol Biotechnol. 2011 Nov;92(3):441-7. doi: 10.1007/s00253-011-3546-2. Epub 2011 Aug 26. Appl Microbiol Biotechnol. 2011. PMID: 21870044 Review.
Cited by
-
Synergistic pathogenesis: exploring biofilms, efflux pumps and secretion systems in Acinetobacter baumannii and Staphylococcus aureus.Arch Microbiol. 2025 May 2;207(6):134. doi: 10.1007/s00203-025-04336-w. Arch Microbiol. 2025. PMID: 40314822 Review.
References
-
- Chen Q, Hao N, Zhao L, Yang X, Yuan Y, Zhao Y, Wang F, Qiu Z, He L, Shi K, Liu S. 2022. Comparative functional analysis of malate metabolism genes in Oenococcus oeni and Lactiplantibacillus plantarum at low pH and their roles in acid stress response. Food Res Int 157:111235. doi:10.1016/j.foodres.2022.111235 - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 2024NC2-GJHX-10/Department of Science and Technology of Shaanxi Province
- 2023BCF01027/Science and Technology Department of Ningxia
- 2022BBF02015/Science and Technology Department of Ningxia
- 2022CXGC010605/Department of Science & Technology of Shandong Province
- 32072206/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources

