TEAL-Seq: targeted expression analysis sequencing
- PMID: 40261045
- PMCID: PMC12108068
- DOI: 10.1128/msphere.00984-24
TEAL-Seq: targeted expression analysis sequencing
Abstract
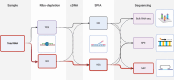
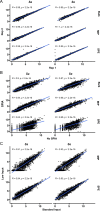
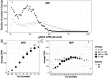
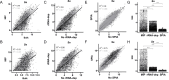
Metagenome sequencing enables the genetic characterization of complex microbial communities. However, determining the activity of isolates within a community presents several challenges, including the wide range of organismal and gene expression abundances, the presence of host RNA, and low microbial biomass at many sites. To address these limitations, we developed "targeted expression analysis sequencing" or TEAL-seq, enabling sensitive species-specific analyses of gene expression using highly multiplexed custom probe pools. For proof of concept, we targeted about 1,700 core and accessory genes of Staphylococcus aureus and S. epidermidis, two key species of the skin microbiome. Two targeting methods were applied to laboratory cultures and human nasal swab specimens. Both methods showed a high degree of specificity, with >90% reads on target, even in the presence of complex microbial or human background DNA/RNA. Targeting using molecular inversion probes demonstrated excellent correlation in inferred expression levels with bulk RNA-seq. Furthermore, we show that a linear pre-amplification step to increase the number of nucleic acids for analysis yielded consistent and predictable results when applied to complex samples and enabled profiling of expression from as little as 1 ng of total RNA. TEAL-seq is much less expensive than bulk metatranscriptomic profiling, enables detection across a greater dynamic range, and uses a strategy that is readily configurable for determining the transcriptional status of organisms in any microbial community.IMPORTANCEThe gene expression patterns of bacteria in microbial communities reflect their activity and interactions with other community members. Measuring gene expression in complex microbiome contexts is challenging, however, due to the large dynamic range of microbial abundances and transcript levels. Here we describe an approach to assessing gene expression for specific species of interest using highly multiplexed pools of targeting probes. We show that an isothermal amplification step enables the profiling of low biomass samples. TEAL-seq should be widely adaptable to the study of microbial activity in natural environments.
Keywords: metagenomics; skin microbiome; targeted sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- van den Berg FF, van Dalen D, Hyoju SK, van Santvoort HC, Besselink MG, Wiersinga WJ, Zaborina O, Boermeester MA, Alverdy J. 2021. Western-type diet influences mortality from necrotising pancreatitis and demonstrates a central role for butyrate. Gut 70:915–927. doi: 10.1136/gutjnl-2019-320430 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

