NEDDylation Regulates CD8+ T-cell Metabolism and Antitumor Immunity
- PMID: 40261130
- PMCID: PMC12214884
- DOI: 10.1158/2326-6066.CIR-24-0127
NEDDylation Regulates CD8+ T-cell Metabolism and Antitumor Immunity
Abstract
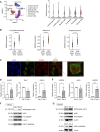
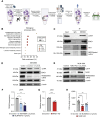
NEDDylation is a posttranslational modification whereby the ubiquitin-like molecule NEDD8 is attached to protein substrates in a process dependent on NEDD8-activating enzyme regulatory subunit (NAE1). NEDDylation is emerging as a regulator of cancer biology, but its precise role in antitumor immunity has not been thoroughly characterized. In this study, we examine the impact of NEDDylation in CD8+ T cell-mediated antitumor responses. Analysis of publicly available single-cell RNA sequencing databases revealed that CD8+ tumor-infiltrating lymphocytes showed increased expression of NEDD8 during their differentiation into effector memory cells. In vitro activation of mouse and human CD8+ T cells drove the upregulation of the NEDDylation enzymatic pathway, resulting in an enrichment of NEDDylated proteins. In vivo tumor challenge assays demonstrated that CD8+ T cells lacking NAE1 exhibited reduced antitumor capability and a less activated phenotype with compromised differentiation into effector cells. Upregulating NEDDylation by knocking out deNEDDylase sentrin-specific protease 8 increased the in vitro cytotoxic capability of CD8+ CAR T cells. In addition, LC MS/MS proteomic analyses of NAE1-deficient CD8+ T cells and CD8+ T cells treated with the NEDDylation inhibitor MLN4924 showed a pronounced impairment in metabolic pathways, including glycolysis and oxidative phosphorylation. In this context, we validated lactate dehydrogenase A, α-enolase, and hexokinase 1, which are relevant glycolytic enzymes, as NEDD8 targets. In line with this, NEDDylation-deficient CD8+ T cells demonstrated reduced transcription, protein expression, and enzymatic activity of lactate dehydrogenase. In summary, we uncover NEDDylation as a critical regulator of CD8+ T cell-mediated antitumor immunity.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
B. Jiménez-Lasheras reports a patent for EP21382780 pending. A. Palazon reports a patent for EP21382780 pending. No disclosures were reported by the other authors.
Figures



References
-
- Sun Y, Wei W, Jin J. Cullin-RING ligases and protein neddylation. Singapore: Springer; 2020. VIII. p. 372.
MeSH terms
Substances
Grants and funding
- PID2019-107956RA-I00/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- PDC2022-133300-I00/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- TED2021-129433B-C21/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- PID2022-139344OB-I00/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- RYC2018-024183-I/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- JDC2022-048612-I/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- PRE2020-092342/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- RYC2021-031213-I/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- PRE2022-104817/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- PID2023-146933OB-100/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- CEX2021-001136-S/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- 804236/HORIZON EUROPE European Research Council (ERC)
- 786462/HORIZON EUROPE European Research Council (ERC)
- PRDVZ21640DEBL/Fundación Científica Asociación Española Contra el Cáncer (AECC)
- PRDVZ1900BOSCH/Fundación Científica Asociación Española Contra el Cáncer (AECC)
- LABAE211744PALA/Fundación Científica Asociación Española Contra el Cáncer (AECC)
- HR21-00925/"la Caixa" Foundation ("la Caixa")
- LCF/BQ/DR20/11790022/"la Caixa" Foundation ("la Caixa")
- PRE_2019_1_0320/Eusko Jaurlaritza (Gobierno Vasco)
- 2023333027/Eusko Jaurlaritza (Gobierno Vasco)
- AF-W1-2019-00012/Eusko Jaurlaritza (Gobierno Vasco)
- XVI fellowship/Fundación Fero (Fundació Fero)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

