Antimicrobial peptide developed with machine learning sequence optimization targets drug resistant Staphylococcus aureus in mice
- PMID: 40261713
- PMCID: PMC12165799
- DOI: 10.1172/JCI185430
Antimicrobial peptide developed with machine learning sequence optimization targets drug resistant Staphylococcus aureus in mice
Abstract
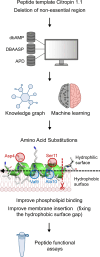
As antimicrobial resistance rises, new antibacterial candidates are urgently needed. Using sequence space information from over 14,743 functional antimicrobial peptides (AMPs), we improved the antimicrobial properties of citropin 1.1, an AMP with weak antimethicillin resistant Staphylococcus aureus (MRSA) activity, producing a short and potent antistaphylococcal peptide, CIT-8 (13 residues). At 40 μg/mL, CIT-8 eradicated 1 × 108 drug-resistant MRSA and vancomycin resistant S. aureus (VRSA) persister cells within 30 minutes of exposure and reduced the number of viable biofilm cells of MRSA and VRSA by 3 log10 and 4 log10 in established biofilms, respectively. CIT-8 (at 32 μg/mL) depolarized and permeated the S. aureus MW2 membrane. In a mouse model of MRSA skin infection, CIT-8 (2% w/w in petroleum jelly) significantly reduced the bacterial burden by 2.3 log10 (P < 0.0001). Our methodology accelerated AMP design by combining traditional peptide design strategies, such as truncation, substitution, and structure-guided alteration, with machine learning-backed sequence optimization.
Keywords: Bacterial infections; Infectious disease; Microbiology.
Conflict of interest statement
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

