Chameleon sequences-Structural effects
- PMID: 40261918
- PMCID: PMC12013887
- DOI: 10.1371/journal.pone.0315901
Chameleon sequences-Structural effects
Abstract
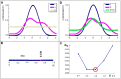
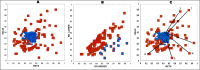
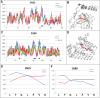
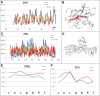
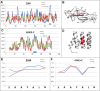
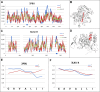
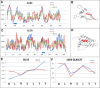
The predisposition of amino acids towards accepting the appropriate secondary structure form is ambiguous. The identified sequences (6-12 aa in length - ChSeq data base) of the chameleon type (the same sequence accepting different secondary structures) constitute a puzzle that makes it difficult to indicate the initial conformation in a chain with a given amino acid sequence. The analysis of proteins presented in this paper uses the hydrophobicity distribution in protein body as the criterion for comaparable analysis of the status of helica/Beta-structural chameleon fragments in pairs of proteins. The sub-base is the object of analysis containg the proteins representing the organisation of hydrophobicity in one protein of the pair as ordered according to micelle-like organisation (hydrophobic core with polar surface) and the second one in pair with disordered hydrophobicity organisation. The status of chameleon sections appears to represent local organisation of hydrophobicity highly accordant in both proteins in chameleon pair independently on the status of the structural unit they belong to. The fuzzy oil drop model (FOD) in its modified form (FOD-M) is applied for analysis. This work aims to verify the hypothesis assuming the subordination of the form of secondary structure to the superior goal of obtaining a hydrophobicity distribution suitable for the given biological activity of the protein, ensuring biological functionality. Secondary structure is not an aim by itself. It is shown, that the main goal is to reach the structure representing specific activity. Secondary structure is a means to achieve this goal.
Copyright: © 2025 Slupina et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare that No competing interests exist.
Figures



References
-
- Hatton CN. LA84 Foundation Digital Library. American Journalism. 2025;42(1):102–3. doi: 10.1080/08821127.2025.2444376 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

