Improved Prediction of CYP2D6 Catalyzed Drug Metabolism by Taking Variant Substrate Specificities and Novel Polymorphic Haplotypes into Account
- PMID: 40261922
- PMCID: PMC12166262
- DOI: 10.1002/cpt.3680
Improved Prediction of CYP2D6 Catalyzed Drug Metabolism by Taking Variant Substrate Specificities and Novel Polymorphic Haplotypes into Account
Abstract
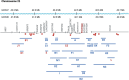
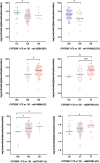
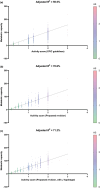
The polymorphic CYP2D6 enzyme plays a pivotal role in the metabolism of approximately 25% of clinically prescribed drugs. However, the impact of specific genetic variants on the interindividual variability in CYP2D6-mediated drug metabolism remains insufficiently quantified. This translational study sought to address this gap by analyzing the genotypes and phenotypes of patients in two large clinical cohorts, focusing on the metabolism of the CYP2D6 substrates risperidone and desmethyltamoxifen. The analysis incorporated novel polymorphic haplotypes and substrate-specific differences among the CYP2D6.1, CYP2D6.2, and CYP2D6.35 enzyme variants. The study revealed that CYP2D6.2 and CYP2D6.35 exhibit reduced metabolic capacity for these substrates, both in vivo and in an in vitro expression model. This was evidenced by decreased catalytic turnover (Kcat), decreased substrate affinity, and altered substrate docking. Furthermore, novel polymorphic haplotypes on the CYP2D6*1, CYP2D6*2, and CYP2D6*35 backgrounds were identified, each associated with a 30-40% increase in CYP2D6 activity. Incorporating these findings into prediction equations significantly improved the genetic prediction accuracy (R2) for CYP2D6-mediated metabolism of desmethyltamoxifen from 59% to 71% and risperidone, also metabolized by CYP3A4, from 42% to 46%. These results highlight the importance of accounting for drug-specific interactions with enzyme variants and integrating distinct polymorphic haplotypes into CYP pharmacogenomic models and guidelines for better translation into clinical practice.
© 2025 The Author(s). Clinical Pharmacology & Therapeutics published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
Magnus Ingelman‐Sundberg, Co‐founder and shareholder in HepaPredict AB. All other authors declared no competing interests for this work.
Figures



References
-
- Table of Pharmacogenetic Associations . <https://www.fda.gov/medical‐devices/precision‐medicine/table‐pharmacogen...>.
-
- Matthaei, J. et al. Heritability of metoprolol and torsemide pharmacokinetics. Clin. Pharmacol. Ther. 98, 611–621 (2015). - PubMed
-
- Ingelman‐Sundberg, M. Pharmacogenomic prescribing guidelines: are they always useful? Clin. Pharmacol. Ther. 0, 1–3 (2024). - PubMed
-
- Jukic, M. , Milosavljević, F. , Molden, E. & Ingelman‐Sundberg, M. Pharmacogenomics in treatment of depression and psychosis: an update. Trends Pharmacol. Sci. 43, 1055–1069 (2022). - PubMed
-
- Zanger, U.M. & Schwab, M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138, 103–141 (2013). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

