NAT10 exacerbates acute renal inflammation by enhancing N4-acetylcytidine modification of the CCL2/CXCL1 axis
- PMID: 40261924
- PMCID: PMC12054813
- DOI: 10.1073/pnas.2418409122
NAT10 exacerbates acute renal inflammation by enhancing N4-acetylcytidine modification of the CCL2/CXCL1 axis
Abstract
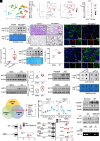
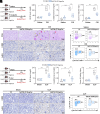
Inflammation plays an essential role in eliminating microbial pathogens and repairing tissues, while sustained inflammation accelerates kidney damage and disease progression. Therefore, understanding the mechanisms of the inflammatory response is vital for developing therapies for inflammatory kidney diseases like acute kidney injury (AKI), which currently lacks effective treatment. Here, we identified N-acetyltransferase 10 (NAT10) as an important regulator for acute inflammation. NAT10, the only known "writer" protein for N4-acetylcytidine (ac4C) acetylation, is elevated in renal tubules across various AKI models, human biopsies, and cultured tubular epithelial cells (TECs). Conditional knockout (cKO) of NAT10 in mouse kidneys attenuates renal dysfunction, inflammation, and infiltration of macrophages and neutrophils, whereas its conditional knock-in (cKI) exacerbates these effects. Mechanistically, our findings from ac4C-RIP-seq and RNA-seq analyses revealed that NAT10-mediated ac4C acetylation enhances the mRNA stability of a range of key chemokines, including C-C motif chemokine ligand 2 (CCL2) and C-X-C motif chemokine ligand 1(CXCL1), promoting macrophage and neutrophil recruitment and accelerating renal inflammation. Additionally, CCL2 and CXCL1 neutralizing antibodies or their receptor inhibitors, abrogated renal inflammation in NAT10-overexpression TECs or NAT10-cKI mice. Importantly, inhibiting NAT10, either through Adeno-associated virus 9 (AAV9)-mediated silencing or pharmacologically with our found inhibitor Cpd-155, significantly reduces renal inflammation and injury. Thus, targeting the NAT10/CCL2/CXCL1 axis presents a promising therapeutic strategy for treating inflammatory kidney diseases.
Keywords: CCL2; CXCL1; NAT10; ac4C; renal inflammation.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures






References
-
- Ronco C., Bellomo R., Kellum J. A., Acute kidney injury. Lancet 394, 1949–1964 (2019). - PubMed
-
- Haines R. W., Fowler A. J., Kirwan C. J., Prowle J. R., The incidence and associations of acute kidney injury in trauma patients admitted to critical care: A systematic review and meta-analysis. J. Trauma Acute Care Surg. 86, 141–147 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

