Quantifying prevalence and risk factors of HIV multiple infection in Uganda from population-based deep-sequence data
- PMID: 40262080
- PMCID: PMC12055032
- DOI: 10.1371/journal.ppat.1013065
Quantifying prevalence and risk factors of HIV multiple infection in Uganda from population-based deep-sequence data
Abstract
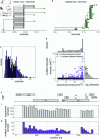
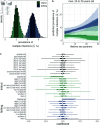
People living with HIV can acquire secondary infections through a process called superinfection, giving rise to simultaneous infection with genetically distinct variants (multiple infection). Multiple infection provides the necessary conditions for the generation of novel recombinant forms of HIV and may worsen clinical outcomes and increase the rate of transmission to HIV seronegative sexual partners. To date, studies of HIV multiple infection have relied on insensitive bulk-sequencing, labor intensive single genome amplification protocols, or deep-sequencing of short genome regions. Here, we identified multiple infections in whole-genome or near whole-genome HIV RNA deep-sequence data generated from plasma samples of 2,029 people living with viremic HIV who participated in the population-based Rakai Community Cohort Study (RCCS). We estimated individual- and population-level probabilities of being multiply infected and assessed epidemiological risk factors using the novel Bayesian deep-phylogenetic multiple infection model (deep - phyloMI) which accounts for bias due to partial sequencing success and false-negative and false-positive detection rates. We estimated that between 2010 and 2020, 4.09% (95% highest posterior density interval (HPD) 2.95%-5.45%) of RCCS participants with viremic HIV multiple infection at time of sampling. Participants living in high-HIV prevalence communities along Lake Victoria were 2.33-fold (95% HPD 1.3-3.7) more likely to harbor a multiple infection compared to individuals in lower prevalence neighboring communities. This work introduces a high-throughput surveillance framework for identifying people with multiple HIV infections and quantifying population-level prevalence and risk factors of multiple infection for clinical and epidemiological investigations.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
Substances
Grants and funding
- D43 TW009578/TW/FIC NIH HHS/United States
- U01 AI100031/AI/NIAID NIH HHS/United States
- R01 HD050180/HD/NICHD NIH HHS/United States
- ZIA AI001040/ImNIH/Intramural NIH HHS/United States
- U01 AI075115/AI/NIAID NIH HHS/United States
- INV-035619/GATES/Gates Foundation/United States
- K25 AI114461/AI/NIAID NIH HHS/United States
- R01 AI155080/AI/NIAID NIH HHS/United States
- R01 HL152813/HL/NHLBI NIH HHS/United States
- R01 HD091003/HD/NICHD NIH HHS/United States
- D43 TW010557/TW/FIC NIH HHS/United States
- R01 AI114438/AI/NIAID NIH HHS/United States
- P30 AI094189/AI/NIAID NIH HHS/United States
- R21 AI145682/AI/NIAID NIH HHS/United States
- R01 AI123002/AI/NIAID NIH HHS/United States
- K01 AI125086/AI/NIAID NIH HHS/United States
- R01 AI143333/AI/NIAID NIH HHS/United States
- R01 AI087409/AI/NIAID NIH HHS/United States
- R01 HD070769/HD/NICHD NIH HHS/United States
- R01 AI128779/AI/NIAID NIH HHS/United States
- R01 AI110324/AI/NIAID NIH HHS/United States
- INV-007573/GATES/Gates Foundation/United States
- L30 AI178824/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

