A network-based analysis to identify a piRNA-target signature related to colorectal cancer prognosis: in silico and in vitro study
- PMID: 40263143
- PMCID: PMC12014994
- DOI: 10.1007/s12672-025-02373-x
A network-based analysis to identify a piRNA-target signature related to colorectal cancer prognosis: in silico and in vitro study
Abstract
Purpose: Patients with colorectal cancer (CRC) are diagnosed in advanced stages and have worse overall survival. Also, this cancer incidence is rising in many countries. The aim of this study is to find piwi-interacting RNAs (piRNA) predicting the prognosis of patients with colorectal cancer, using bioinformatics and evaluating these results through RT-qPCR method.
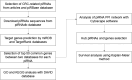
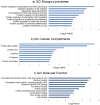
Methods: The target genes of piRNAs were predicted using miRDB and TargetRank databases. Protein-protein interaction (PPI) networks were constructed by STRING and were analyzed with Cytoscape software and the MCODE tool used for module construction. Expression levels of final selected piRNAs in 18 pairs of CRC tissue and adjacent normal tissue were measured by quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR).
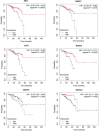
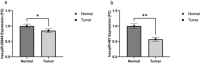
Results: Twenty CRC-related piRNAs and 980 target genes were included in this study. After PPI analysis 19 hub genes were identified. Then, the prognostic value of these hub genes was assessed via Kaplan-Meier survival analyses. This survival analysis indicated that the expression of six genes was significantly associated with overall survival of patients with CRC. These genes are targets of hsa-piR-487, hsa-piR-28944 and piR-hsa-8401. Also, the pathway analysis revealed the potential signal pathways of these piRNAs targets involved in CRC. RT-qPCR showed that hsa-piR-487 and hsa-piR-28944 expression significantly were down-regulated in CRC tumor tissues compared with the adjacent normal tissues (P < 0.05, P < 0.01).
Conclusion: It seems that hsa-piR-487 and hsa-piR-28944 can be considered as a potential biomarker for the diagnosis of CRC. However, it is still necessary to conduct studies with a higher statistical population and measure them in the serum of patients to confirm these results.
Keywords: Biomarkers; Colorectal neoplasms; Piwi-interacting RNA; Prognosis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the Ethics Committee of Hamadan University of Medical Sciences (IR.UMSHA.REC.1400.879). Informed consent was obtained from the participants to participate in the current study (in Persian). Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
PIWI-interacting RNAs are aberrantly expressed and may serve as novel biomarkers for diagnosis of lung adenocarcinoma.Thorac Cancer. 2021 Sep;12(18):2468-2477. doi: 10.1111/1759-7714.14094. Epub 2021 Aug 3. Thorac Cancer. 2021. PMID: 34346164 Free PMC article.
-
Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer.Mol Cancer. 2018 Jan 30;17(1):16. doi: 10.1186/s12943-018-0767-3. Mol Cancer. 2018. PMID: 29382334 Free PMC article.
-
A serum piRNA signature as promising non-invasive diagnostic and prognostic biomarkers for colorectal cancer.Cancer Manag Res. 2019 Apr 29;11:3703-3720. doi: 10.2147/CMAR.S193266. eCollection 2019. Cancer Manag Res. 2019. PMID: 31118791 Free PMC article.
-
Piwi-Interacting RNAs: A New Class of Regulator in Human Breast Cancer.Front Oncol. 2021 Jul 6;11:695077. doi: 10.3389/fonc.2021.695077. eCollection 2021. Front Oncol. 2021. PMID: 34295823 Free PMC article. Review.
-
The potential of piR-823 as a diagnostic biomarker in oncology: A systematic review.PLoS One. 2023 Dec 7;18(12):e0294685. doi: 10.1371/journal.pone.0294685. eCollection 2023. PLoS One. 2023. PMID: 38060527 Free PMC article.
References
-
- Hassan C, et al. Meta-analysis: adherence to colorectal cancer screening and the detection rate for advanced neoplasia, according to the type of screening test. Aliment Pharmacol Ther. 2012;36(10):929–40. 10.1111/apt.12071. - PubMed
-
- Jing Z, et al. Biological roles of piRNAs in colorectal cancer. Gene. 2021;769: 145063. 10.1016/j.gene.2020.145063. - PubMed
LinkOut - more resources
Full Text Sources
