Make-or-break prime editing for genome engineering in Streptococcus pneumoniae
- PMID: 40263274
- PMCID: PMC12015366
- DOI: 10.1038/s41467-025-59068-8
Make-or-break prime editing for genome engineering in Streptococcus pneumoniae
Abstract
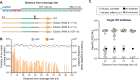
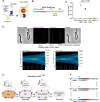
CRISPR-Cas9 has revolutionized genome engineering by allowing precise introductions of DNA double-strand breaks (DSBs). However, genome engineering in bacteria is still a complex, multi-step process requiring a donor DNA template for repair of DSBs. Prime editing circumvents this need as the repair template is indirectly provided within the prime editing guide RNA (pegRNA). Here, we developed make-or-break Prime Editing (mbPE) that allows for precise and effective genetic engineering in the opportunistic human pathogen Streptococcus pneumoniae. In contrast to traditional prime editing in which a nicking Cas9 is employed, mbPE harnesses wild type Cas9 in combination with a pegRNA that destroys the seed region or protospacer adjacent motif. Since most bacteria poorly perform template-independent end joining, correctly genome-edited clones are selectively enriched during mbPE. We show that mbPE is RecA-independent and can be used to introduce point mutations, deletions and targeted insertions, including protein tags such as a split luciferase, at selection efficiencies of over 93%. mbPE enables sequential genome editing, is scalable, and can be used to generate pools of mutants in a high-throughput manner. The mbPE system and pegRNA design guidelines described here will ameliorate future bacterial genome editing endeavors.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.W.V. is a scientific advisory board member at i-Seq Biotechnology. The remaining authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- 310030_200792/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- NCCR AntiResist 51NF40_180541/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- TMPFP3_210202/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- 771534-PneumoCaTChER/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources

