Multi-omics profiling of dairy cattle oxidative stress identifies hindgut-derived Phascolarctobacterium succinatutens exhibiting antioxidant activity
- PMID: 40263287
- PMCID: PMC12015594
- DOI: 10.1038/s41522-025-00698-7
Multi-omics profiling of dairy cattle oxidative stress identifies hindgut-derived Phascolarctobacterium succinatutens exhibiting antioxidant activity
Abstract
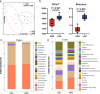
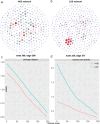
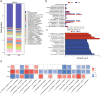
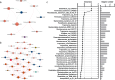
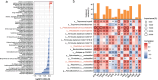
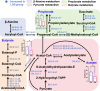
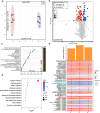
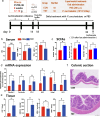
An imbalance between oxidative and antioxidant processes in the host can lead to excessive oxidation, a condition known as oxidative stress (OS). Although changes in the hindgut microbiota have been frequently linked to OS, the specific microbial and metabolic underpinnings of this association remain unclear. In this study, we enrolled 81 postpartum Holstein cows and stratified them into high oxidative stress (HOS, n = 9) and low oxidative stress (LOS, n = 9) groups based on the oxidative stress index (OSi). Using a multi-omics approach, we performed 16S rRNA gene sequencing to evaluate microbial diversity, conducted metagenomic analysis to identify functional bacteria, and utilized untargeted metabolomics to profile serum metabolites. Our analyses revealed elevated levels of kynurenine, formyl-5-hydroxykynurenamine, and 5-hydroxyindole-3-acetic acid in LOS dairy cows. Additionally, the LOS cows had a higher abundance of short-chain fatty acids (SCFAs)-producing bacteria, including Bacteroidetes bacterium, Paludibacter propionicigenes, and Phascolarctobacterium succinatutens (P. succinatutens), which were negatively correlated with OSi. To explore the potential role of these bacteria in mitigating OS, we administered P. succinatutens (108 cfu/day for 14 days) to C57BL/6 J mice (n = 10). Oral administration of P. succinatutens significantly increased serum total antioxidant capacity, decreased total oxidants, and reduced OSi in mice. Moreover, this treatment promoted activation of the Nrf2-Keap1 antioxidant pathway, significantly enhancing the enzymatic activities of GSH-Px and SOD, as well as the concentrations of acetate and propionate in the colon. In conclusion, our findings suggest that systemic tryptophan metabolism and disordered SCFAs production are concurrent factors influenced by hindgut microbiota and associated with OS development. Modulating the hindgut microbiota, particularly by introducing specific SCFAs-producing bacteria, could be a promising strategy for combating OS.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: The animal care protocol received approval from the Animal Care and Use Committee of China Agricultural University (Protocol Number: AW10803202-3-1) and the Beijing Laboratory Animal Research Center (Protocol Number: BLARC-LAWER-202404003).
Figures
Similar articles
-
The hindgut microbiome contributes to host oxidative stress in postpartum dairy cows by affecting glutathione synthesis process.Microbiome. 2023 Apr 22;11(1):87. doi: 10.1186/s40168-023-01535-9. Microbiome. 2023. PMID: 37087457 Free PMC article.
-
Integrated multi-omics analysis reveals the positive leverage of citrus flavonoids on hindgut microbiota and host homeostasis by modulating sphingolipid metabolism in mid-lactation dairy cows consuming a high-starch diet.Microbiome. 2023 Oct 25;11(1):236. doi: 10.1186/s40168-023-01661-4. Microbiome. 2023. PMID: 37880759 Free PMC article.
-
Live yeast supplementation altered the bacterial community's composition and function in rumen and hindgut and alleviated the detrimental effects of heat stress on dairy cows.J Anim Sci. 2023 Jan 3;101:skac410. doi: 10.1093/jas/skac410. J Anim Sci. 2023. PMID: 36534956 Free PMC article.
-
Rumen and hindgut microbiome regulate average daily gain of preweaning Holstein heifer calves in different ways.Microbiome. 2024 Jul 19;12(1):131. doi: 10.1186/s40168-024-01844-7. Microbiome. 2024. PMID: 39030599 Free PMC article.
-
Imbalance of Microbacterial Diversity Is Associated with Functional Prognosis of Stroke.Neural Plast. 2023 May 8;2023:6297653. doi: 10.1155/2023/6297653. eCollection 2023. Neural Plast. 2023. PMID: 37197229 Free PMC article.
Cited by
-
Rutin Attenuates Oxidative Stress Responses and Hepatocyte Metabolomics in β-Hydroxybutyric Acid-Induced Hepatocyte Injury in Calves.Int J Mol Sci. 2025 Jun 19;26(12):5878. doi: 10.3390/ijms26125878. Int J Mol Sci. 2025. PMID: 40565341 Free PMC article.
References
-
- Abuelo, A., Hernández, J., Benedito, J. L. & Castillo, C. The importance of the oxidative status of dairy cattle in the periparturient period: revisiting antioxidant supplementation. J. Anim. Physiol. Anim. Nutr.99, 1003–1016 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

