Antibody function predicts viral control in newborn monkeys immunised with an influenza virus HA stem nanoparticle
- PMID: 40263387
- PMCID: PMC12015251
- DOI: 10.1038/s41467-025-59149-8
Antibody function predicts viral control in newborn monkeys immunised with an influenza virus HA stem nanoparticle
Abstract
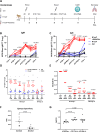
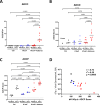
The lack of an approved influenza vaccine for infants <6 months, coupled with the requirement for annual updates of current vaccines, warrants the development of a universal vaccine that can confer protection in young infants. Here we test the ability of a ferritin nanoparticle universal influenza vaccine (H1ssF) containing the stem region of hemagglutinin (HA) adjuvanted with AddaVax to elicit responses in newborn African green monkeys (AGM). Vaccinated newborns show robust HA stem-specific IgG responses but, despite the high antibody levels, viral load in the lung following H1N1 Ca09 challenge is variable among animals. Further analysis indicates that viral clearance is correlated with the presence of antibodies with neutralizing and antibody-dependent cellular phagocytosis activity. Our findings show that newborn AGM can generate functional HA stem-specific antibodies for viral clearance following vaccination with H1ssF+AddaVax and support further investigation of H1ssF as a universal vaccine for this vulnerable human population.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.K. is listed as inventor of patents and patent applications on vaccine immunogens used in this study filed by the U.S. Department of Health and Human Services. The remaining authors declare no competing interests.
Figures




References
-
- Esposito, S. & Principi, N. The rational use of influenza vaccines in healthy children and children with underlying conditions. Curr. Opin. Infect. Dis.22, 244–249 (2009). - PubMed
-
- Levy, O. Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat. Rev. Immunol.7, 379–390 (2007). - PubMed
MeSH terms
Substances
Grants and funding
- P40 OD010965/OD/NIH HHS/United States
- T32AI007401/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- P30 CA012197/CA/NCI NIH HHS/United States
- R01AI146059/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- P30CA012197/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical

