Lipoprotein Lpp and L, D-transpeptidases regulate the master regulator of virulence AggR in EAEC
- PMID: 40263412
- PMCID: PMC12015436
- DOI: 10.1038/s41598-025-96373-0
Lipoprotein Lpp and L, D-transpeptidases regulate the master regulator of virulence AggR in EAEC
Abstract
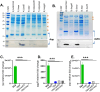
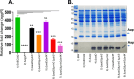
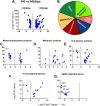
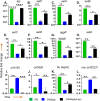
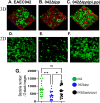
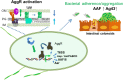
Enteroaggregative Escherichia coli (EAEC) is a diarrheagenic pathotype associated with traveler's diarrhea, foodborne outbreaks, and sporadic diarrhea in industrialized and developing countries. Regulation of virulence factors in EAEC is mediated by the master regulator AggR, an AraC/XylS family member controlling the expression of more than 44 genes associated with metabolism and virulence. Although the AggR regulon is well-characterized, the mechanism and upstream signaling cascades that regulate its activation are poorly understood. This study demonstrates that Lpp (Braun's lipoprotein) and L, D-transpeptidases are required for AggR activation. We found that deletion lpp in EAEC resulted in the downregulation of more than 100 genes involved in transport, metabolism, and virulence. Among the genes, fourteen transcriptional factors, including AggR, were differentially expressed in 042Δlpp. Our findings also showed that Lpp anchoring to the peptidoglycan is a requisite for AggR-activation. Hence, chemical inhibition or genetic deletion of L, D-transpeptidases encoding genes involved in the crosslink of Lpp to the peptidoglycan abolished AggR activation. Moreover, the 042Δlpp mutant exhibited reduced biofilm formation on abiotic surfaces and reduced colonization of human intestinal colonoids. This is the first study to demonstrate the tight regulation of the AraC/XylS transcriptional regulator AggR, essential in EAEC virulence and intestinal colonization by components of the bacterial cell envelope.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries.J Clin Microbiol. 2007 Jan;45(1):121-6. doi: 10.1128/JCM.01128-06. Epub 2006 Nov 8. J Clin Microbiol. 2007. PMID: 17093030 Free PMC article.
-
Enteroaggregative Escherichia coli (EAEC) isolates obtained from non-diarrheic children carry virulence factor-encoding genes from Extraintestinal Pathogenic E. Coli (ExPEC).Braz J Microbiol. 2024 Dec;55(4):3551-3561. doi: 10.1007/s42770-024-01471-2. Epub 2024 Jul 31. Braz J Microbiol. 2024. PMID: 39083223 Free PMC article.
-
Dual Function of Aar, a Member of the New AraC Negative Regulator Family, in Escherichia coli Gene Expression.Infect Immun. 2020 May 20;88(6):e00100-20. doi: 10.1128/IAI.00100-20. Print 2020 May 20. Infect Immun. 2020. PMID: 32253248 Free PMC article.
-
A Mini-Review of Enteroaggregative Escherichia coli with a Specific Target on the Virulence Factors Controlled by the AggR Master Regulator.Pol J Microbiol. 2023 Dec 16;72(4):347-354. doi: 10.33073/pjm-2023-037. eCollection 2023 Dec 1. Pol J Microbiol. 2023. PMID: 37875068 Free PMC article. Review.
-
Prevalence of specific serogroups, antibiotic resistance and virulence factors of avian pathogenic Escherichia coli (APEC) isolated from clinical cases: A systematic review and meta-analysis.Microb Pathog. 2024 Sep;194:106843. doi: 10.1016/j.micpath.2024.106843. Epub 2024 Aug 6. Microb Pathog. 2024. PMID: 39117015
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

