Genomic surveillance of SARS-CoV-2 variants using pooled WGS
- PMID: 40263437
- PMCID: PMC12015266
- DOI: 10.1038/s41598-025-99201-7
Genomic surveillance of SARS-CoV-2 variants using pooled WGS
Abstract
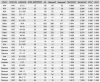
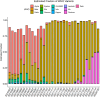
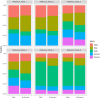
This study presents the development and validation of a genomic surveillance strategy using Whole Genome Sequencing (WGS) on normalized pooled samples to detect and monitor SARS-CoV-2 variants. A bioinformatics pipeline was designed specifically for analyzing pooled WGS data and was validated using simulated datasets, pooled samples of reference materials, and pooled clinical samples collected during key periods of the Delta and Omicron variant emergence. The approach was evaluated for its accuracy in estimating variant abundance at both the Phylogenetic Assignment of Named Global Outbreak (PANGO) lineage level and the World Health Organization (WHO) variant level. From the simulation datasets, the method achieved an overall sensitivity of 99.1% and a positive predictive value (PPV) of 99.9% for detecting SARS-CoV-2 variants at the WHO variant level. At the PANGO lineage level, it achieved an overall sensitivity of 82.8% and a PPV of 77.4% when a predicted lineage was considered accurate if it shared more than 90% of markers with any true lineage present in the pooled sample. The accuracy of variant abundance estimation was further validated using pooled samples of reference materials. Analysis of pooled clinical samples showed results consistent with national epidemiological trends, particularly during the emergence of the Delta and Omicron variants in Korea. This pooled WGS-based genomic surveillance strategy offers a scalable and economical solution for monitoring SARS-CoV-2 variants, providing public health authorities with a valuable tool for tracking pandemic dynamics and enabling timely responses.
Keywords: Epidemiological surveillance; SARS-CoV-2; SARS-CoV-2 mutation screening.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical statement: The study was approved by the Institutional Review Board of Gangnam Severance Hospital (IRB number 3-2022-0459), and adhered to the clinical practice guidelines of the Declaration of Helsinki (2013 amendment).
Figures



Similar articles
-
Prospective clinical performance of CoVarScan in identifying SARS-CoV-2 Omicron subvariants.Microbiol Spectr. 2025 Jan 7;13(1):e0138524. doi: 10.1128/spectrum.01385-24. Epub 2024 Dec 11. Microbiol Spectr. 2025. PMID: 39660915 Free PMC article.
-
Genomics-based approach for detection and characterization of SARS-CoV-2 co-infections and diverse viral populations.Microbiol Spectr. 2025 Jun 3;13(6):e0209224. doi: 10.1128/spectrum.02092-24. Epub 2025 May 1. Microbiol Spectr. 2025. PMID: 40310264 Free PMC article.
-
SARS-CoV-2 introductions to the island of Ireland: a phylogenetic and geospatiotemporal study of infection dynamics.Genome Med. 2024 Dec 19;16(1):150. doi: 10.1186/s13073-024-01409-1. Genome Med. 2024. PMID: 39702217 Free PMC article.
-
Nosocomial Outbreak of SARS-CoV-2 in a Hospital Ward during the Omicron Variant-Dominant Wave with a Review of the Relevant Literature.Jpn J Infect Dis. 2024 Sep 19;77(5):253-259. doi: 10.7883/yoken.JJID.2023.464. Epub 2024 May 31. Jpn J Infect Dis. 2024. PMID: 38825458 Review.
-
Dynamics of SARS-CoV-2 variants in West Africa: Insights into genomic surveillance in resource-constrained settings.Infect Genet Evol. 2024 Nov;125:105681. doi: 10.1016/j.meegid.2024.105681. Epub 2024 Oct 20. Infect Genet Evol. 2024. PMID: 39437881 Free PMC article. Review.
References
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

