Museomics of an extinct European flat oyster population
- PMID: 40263463
- PMCID: PMC12015263
- DOI: 10.1038/s41598-025-96743-8
Museomics of an extinct European flat oyster population
Abstract
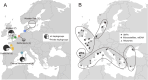
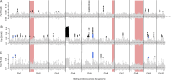
Understanding the factors that predispose species and populations to decline and extinction is a major challenge of biodiversity research. In the present study, we investigated the historical population genomics of an extinct population of the European oyster (Ostrea edulis L.) from the Wadden Sea collected between 1868 and 1888, and compared it to French and English populations sampled at the same time. Our museomic results indicate that the now-extinct population was genetically isolated from the French and English populations and showed signs of local adaptation in the form of Fst outlier loci between the Wadden Sea and the other two populations. Thus the Wadden Sea oysters may have been predisposed for extinction because they were not naturally replenished from other populations. A comparison of population-wide genomic diversity may hint towards a sudden population contraction of the Wadden Sea population, possibly being the result of stronger - or earlier - population decline in this population than in the others. In summary, our historical population genomic exploration hints at some potential causes of population decline in flat oysters from the Wadden Sea, which might have led to their extinction.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Phylogeography in an "oyster" shell provides first insights into the genetic structure of an extinct Ostrea edulis population.Sci Rep. 2021 Jan 27;11(1):2307. doi: 10.1038/s41598-021-82020-x. Sci Rep. 2021. PMID: 33504886 Free PMC article.
-
Coming and going - Historical distributions of the European oyster Ostrea edulis Linnaeus, 1758 and the introduced slipper limpet Crepidula fornicata Linnaeus, 1758 in the North Sea.PLoS One. 2019 Oct 24;14(10):e0224249. doi: 10.1371/journal.pone.0224249. eCollection 2019. PLoS One. 2019. PMID: 31648244 Free PMC article.
-
The late Holocene demise of a sublittoral oyster bed in the North Sea.PLoS One. 2021 Feb 16;16(2):e0242208. doi: 10.1371/journal.pone.0242208. eCollection 2021. PLoS One. 2021. PMID: 33591987 Free PMC article.
-
The future of Baltic Sea populations: local extinction or evolutionary rescue?Ambio. 2011 Mar;40(2):179-90. doi: 10.1007/s13280-010-0129-x. Ambio. 2011. PMID: 21446396 Free PMC article. Review.
-
Understanding local plant extinctions before it is too late: bridging evolutionary genomics with global ecology.New Phytol. 2023 Mar;237(6):2005-2011. doi: 10.1111/nph.18718. Epub 2023 Jan 27. New Phytol. 2023. PMID: 36604850 Review.
References
-
- Biosafety Unit. Status and Trends of Global Biodiversity. in Global Biodiversity Outlook. Secretariat of the Convention on Biological Diversity, Quebec, Canada, (2001).
-
- Cardinale, B. J. et al. Biodiversity loss and its impact on humanity. Nature486, 59–67 (2012). - PubMed
-
- Dirzo, R. et al. Defaunation in the anthropocene. Science345, 401–406 (2014). - PubMed
-
- Ceballos, G. & Ehrlich, P. R. Mammal population losses and the extinction crisis. Science296, 904–907 (2002). - PubMed
-
- Collen, B. et al. Monitoring change in vertebrate abundance: the living planet index. Conserv. Biol.23, 317–327 (2009). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

