Metastatic medulloblastoma remodels the local leptomeningeal microenvironment to promote further metastatic colonization and growth
- PMID: 40263572
- PMCID: PMC12081294
- DOI: 10.1038/s41556-025-01660-7
Metastatic medulloblastoma remodels the local leptomeningeal microenvironment to promote further metastatic colonization and growth
Abstract
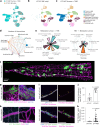
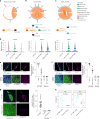
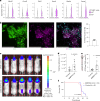
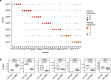
Leptomeningeal metastases are the major source of morbidity and mortality for patients with medulloblastoma. The biology of the leptomeningeal metastases and the local tumour microenvironment are poorly characterized. Here we show that metastasis-associated meningeal fibroblasts (MB-MAFs) are transcriptionally distinct and signal extensively to tumour cells and the tumour microenvironment. Metastatic cells secrete platelet-derived growth factor (PDGF) ligands into the local microenvironment to chemotactically recruit meningeal fibroblasts. Meningeal fibroblasts are reprogrammed to become MB-MAFs, expressing distinct transcriptomes and secretomes, including bone morphogenetic proteins. Active bone morphogenetic protein signalling and co-implantation of tumour cells with MB-MAFs enhances the colonization of the leptomeninges by medulloblastoma cells and promotes the growth of established metastases. Furthermore, treatment of patient-derived xenograft mice with a PDGF-receptor-α neutralizing antibody enhances overall survival in vivo. Collectively, our results define a targetable intercellular communication cascade in the metastatic niche to treat leptomeningeal disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures












References
MeSH terms
Substances
Grants and funding
- R01CA159859/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01CA255369/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- CPRIT - RR220051/Cancer Prevention and Research Institute of Texas (Cancer Prevention Research Institute of Texas)
- SU2C-AACR-DT-19-15/EIF | Stand Up To Cancer (SU2C)
- R01 CA255369/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

