SDCBP/Syntenin-1 stabilizes BACH1 by disassembling the SCFFBXO22-BACH1 complex in triple-negative breast cancer
- PMID: 40263598
- PMCID: PMC12130529
- DOI: 10.1038/s44318-025-00440-1
SDCBP/Syntenin-1 stabilizes BACH1 by disassembling the SCFFBXO22-BACH1 complex in triple-negative breast cancer
Abstract
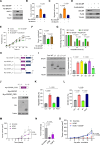
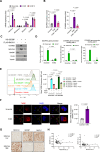
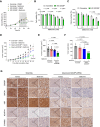
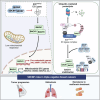
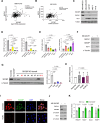
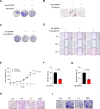
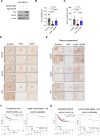
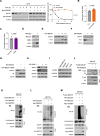
BACH1 is a redox-sensitive transcription factor facilitating tumor progression in triple-negative breast cancer (TNBC). However, the molecular mechanisms regulating BACH1 function in TNBC remain unclear. In this study, we demonstrate that SDCBP, a tandem-PDZ-domain protein, stabilizes BACH1 by disassembling the Skp1-Cullin1-FBXO22 (SCFFBXO22)-BACH1 complex via a heme/heme-oxygenase-1-independent manner in TNBC cells. Our data revealed that SDCBP and BACH1 expression show a significant positive correlation in TNBC cells and TNBC patients tumor tissues. Mechanistically, SDCBP via its PDZ1 domain disassembles the SCFFBXO22-BACH1 complex via its PDZ1 domain, thereby preventing BACH1 K48-linked polyubiquitination and proteasomal degradation. Knocking down SDCBP induces BACH1 degradation and downregulates expressions of BACH1-induced metastatic genes, thereby suppressing tumor progression in mice bearing TNBC tumors. Moreover, depleting SDCBP leads to upregulation of BACH1-repressed electron transport chain (ETC) genes, such as NDUFA4 and COX6B2, and increases mitochondrial activity, enhancing anti-tumor efficacy of metformin against TNBC both in vitro and in vivo. These data demonstrate a novel alternative mechanism for BACH1 stabilization mediated by SDCBP, implicating the SDCBP-BACH1 axis as a potential target for enhancing ETC inhibitor efficacy in TNBC combinational therapy.
Keywords: BACH1; Metformin; SDCBP; Triple-negative Breast Cancer; Ubiquitin E3 Ligase SCFFBXO22.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures



References
-
- Amoutzias GD, Veron AS, Weiner J, Robinson-Rechavi M, Bornberg-Bauer E, Oliver SG, Robertson DL (2007) One billion years of bZIP transcription factor evolution: conservation change in dimerization DNA-binding site specificity. Mol Biol Evol 24:827–835 - PubMed
-
- Copel RL, Kanaan Y (2021) New targets in triple-negative breast cancer. Nat Rev Cancer 21:744 - PubMed
-
- Crowley LC, Christensen ME, Waterhouse NJ (2016) Measuring mitochondrial transmembrane potential by TMRE staining. Cold Spring Harb Protoc 2016:pdb-prot087361 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

