IL-10 targets IRF transcription factors to suppress IFN and inflammatory response genes by epigenetic mechanisms
- PMID: 40263613
- PMCID: PMC12640682
- DOI: 10.1038/s41590-025-02137-3
IL-10 targets IRF transcription factors to suppress IFN and inflammatory response genes by epigenetic mechanisms
Abstract
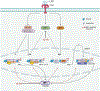
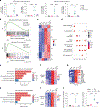
Interleukin-10 (IL-10) is pivotal in suppressing innate immune activation, in large part by suppressing induction of inflammatory genes. Despite decades of research, the molecular mechanisms underlying this inhibition have not been resolved. Here we utilized an integrated epigenomic analysis to investigate IL-10-mediated suppression of LPS and TNF responses in primary human monocytes. Instead of inhibiting core TLR4-activated pathways such as NF-κB, MAPK-AP-1 and TBK1-IRF3 signaling, IL-10 targeted IRF transcription factor activity and DNA binding, particularly IRF5 and an IRF1-mediated amplification loop. This resulted in suppression of inflammatory NF-κB target genes and near-complete suppression of interferon-stimulated genes. Mechanisms of gene inhibition included downregulation of chromatin accessibility, de novo enhancer formation and IRF1-associated H3K27ac activating histone marks. These results provide a mechanism by which IL-10 suppresses inflammatory NF-κB target genes, highlight the role of IRF1 in inflammatory gene expression and describe the suppression of IFN responses by epigenetic mechanisms.
© 2025. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



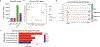








References
-
- Ouyang W & O’Garra A IL-10 family cytokines IL-10 and IL-22: from basic science to clinical translation. Immunity 50, 871–891 (2019). - PubMed
-
- Kuhn R, Lohler J, Rennick D, Rajewsky K & Muller W Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993). - PubMed
-
- Zigmond E et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733 (2014). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

