The retrotransposon-derived capsid genes PNMA1 and PNMA4 maintain reproductive capacity
- PMID: 40263616
- PMCID: PMC12180178
- DOI: 10.1038/s43587-025-00852-y
The retrotransposon-derived capsid genes PNMA1 and PNMA4 maintain reproductive capacity
Abstract
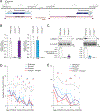
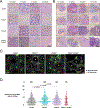
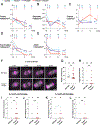
Almost half of the human genome consists of retrotransposons-'parasitic' sequences that insert themselves into the host genome via an RNA intermediate. Although most of these sequences are silenced or mutationally deactivated, they can present opportunities for evolutionary innovation: mutation of a deteriorating retrotransposon can result in a gene that provides a selective advantage to the host in a process termed 'domestication'1-3. The PNMA family of gag-like capsid genes was domesticated from an ancient vertebrate retrotransposon of the Metaviridae clade at least 100 million years ago4,5. PNMA1 and PNMA4 are positively regulated by the master germ cell transcription factors MYBL1 and STRA8, and their transcripts are bound by the translational regulator DAZL during gametogenesis6. This developmental regulation of PNMA1 and PNMA4 expression in gonadal tissue suggested to us that they might serve a reproductive function. Through the analysis of donated human ovaries, genome-wide association studies (GWASs) and mouse models, we found that PNMA1 and PNMA4 are necessary for the maintenance of a normal reproductive lifespan. These proteins self-assemble into capsid-like structures that exit human cells, and we observed large PNMA4 particles in mouse male gonadal tissue that contain RNA and are consistent with capsid formation.
© 2025. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: T.W.P.W., H.B.C. and L.E.B. are co-inventors on a PCT international patent application filed by Columbia University related to this work (PCT/US2024/053329). B.W. is the founder of SurGene, LLC and an inventor on patent applications related to CRISPR–Cas systems and applications thereof. All other authors declare no competing interests.
Figures




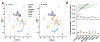




Update of
-
The retrotransposon - derived capsid genes PNMA1 and PNMA4 maintain reproductive capacity.bioRxiv [Preprint]. 2024 May 14:2024.05.11.592987. doi: 10.1101/2024.05.11.592987. bioRxiv. 2024. Update in: Nat Aging. 2025 May;5(5):765-779. doi: 10.1038/s43587-025-00852-y. PMID: 38798495 Free PMC article. Updated. Preprint.
-
The retrotransposon-derived capsid genes PNMA1 and PNMA4 maintain reproductive capacity.Res Sq [Preprint]. 2024 Jul 10:rs.3.rs-4559920. doi: 10.21203/rs.3.rs-4559920/v1. Res Sq. 2024. Update in: Nat Aging. 2025 May;5(5):765-779. doi: 10.1038/s43587-025-00852-y. PMID: 39041030 Free PMC article. Updated. Preprint.
References
-
- Naville M et al. Not so bad after all: retroviruses and long terminal repeat retrotransposons as a source of new genes in vertebrates. Clin. Microbiol. Infect. 22, 312–323 (2016). - PubMed
MeSH terms
Substances
Grants and funding
- R35 GM134867/GM/NIGMS NIH HHS/United States
- NA/M.J. Murdock Charitable Trust (Murdock Charitable Trust)
- R01 AG069750/AG/NIA NIH HHS/United States
- R01 NS109858/NS/NINDS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AG069742/AG/NIA NIH HHS/United States
- R35 GM136340/GM/NIGMS NIH HHS/United States
- 213470/A/18/Z/Wellcome Trust (Wellcome)
- R35 GM146725/GM/NIGMS NIH HHS/United States
- R35 GM124633/GM/NIGMS NIH HHS/United States
- R35 GM147130/GM/NIGMS NIH HHS/United States
- NA/Irma T. Hirschl Trust (Irma T. Hirschl Charitable Trust)
- R35 GM140876/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources

