DGCLCMI: a deep graph collaboration learning method to predict circRNA-miRNA interactions
- PMID: 40264118
- PMCID: PMC12016396
- DOI: 10.1186/s12915-025-02197-9
DGCLCMI: a deep graph collaboration learning method to predict circRNA-miRNA interactions
Abstract
Background: Numerous studies have shown that circRNA can act as a miRNA sponge, competitively binding to miRNAs, thereby regulating gene expression and disease progression. Due to the high cost and time-consuming nature of traditional wet lab experiments, analyzing circRNA-miRNA associations is often inefficient and labor-intensive. Although some computational models have been developed to identify these associations, they fail to capture the deep collaborative features between circRNA and miRNA interactions and do not guide the training of feature extraction networks based on these high-order relationships, leading to poor prediction performance.
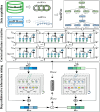
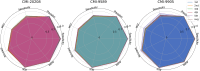
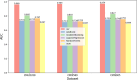
Results: To address these issues, we innovatively propose a novel deep graph collaboration learning method for circRNA-miRNA interaction, called DGCLCMI. First, it uses word2vec to encode sequences into word embeddings. Next, we present a joint model that combines an improved neural graph collaborative filtering method with a feature extraction network for optimization. Deep interaction information is embedded as informative features within the sequence representations for prediction. Comprehensive experiments on three well-established datasets across seven metrics demonstrate that our algorithm significantly outperforms previous models, achieving an average AUC of 0.960. In addition, a case study reveals that 18 out of 20 predicted unknown CMI data points are accurate.
Conclusions: The DGCLCMI improves circRNA and miRNA feature representation by capturing deep collaborative information, achieving superior performance compared to prior methods. It facilitates the discovery of unknown associations and sheds light on their roles in physiological processes.
Keywords: CircRNA-miRNA interaction; Collaborative filtering; Graph neural networks; LSTM; Word2vec.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

