The 15-Year Survival Advantage: Immune Resilience as a Salutogenic Force in Healthy Aging
- PMID: 40264357
- PMCID: PMC12266754
- DOI: 10.1111/acel.70063
The 15-Year Survival Advantage: Immune Resilience as a Salutogenic Force in Healthy Aging
Abstract
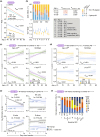
Human aging presents an evolutionary paradox: while aging rates remain constant, healthspan and lifespan vary widely. We address this conundrum via salutogenesis-the active production of health-through immune resilience (IR), the capacity to resist disease despite aging and inflammation. Analyzing ~17,500 individuals across lifespan stages and inflammatory challenges, we identified a core salutogenic mechanism: IR centered on TCF7, a conserved transcription factor maintaining T-cell stemness and regenerative potential. IR integrates innate and adaptive immunity to counter three aging and mortality drivers: chronic inflammation (inflammaging), immune aging, and cellular senescence. By mitigating these aging mechanisms, IR confers survival advantages: At age 40, individuals with poor IR face a 9.7-fold higher mortality rate-a risk equivalent to that of 55.5-year-olds with optimal IR-resulting in a 15.5-year gap in survival. Optimal IR preserves youthful immune profiles at any age, enhances vaccine responses, and reduces burdens of cardiovascular disease, Alzheimer's, and serious infections. Two key salutogenic evolutionary themes emerge: first, female-predominant IR, including TCF7, likely reflects evolutionary pressures favoring reproductive success and caregiving; second, midlife (40-70 years) is a critical window where optimal IR reduces mortality by 69%. After age 70, mortality rates converge between resilient and non-resilient groups, reflecting biological limits on longevity extension. TNFα-blockers restore salutogenesis pathways, indicating IR delays aging-related processes rather than altering aging rates. By reframing aging as a salutogenic-pathogenic balance, we establish TCF7-centered IR as central to healthy longevity. Targeted midlife interventions to enhance IR offer actionable strategies to maximize healthspan before biological constraints limit benefits.
Keywords: Alzheimer's disease; T cell; accelerated aging; cardiac declines with age; inflammation; lifespan; longevity regulation; senescence.
Published 2025. This article is a U.S. Government work and is in the public domain in the USA. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








References
-
- Azambuja, M. I. 2004. “Spanish Flu and Early 20th‐Century Expansion of a Coronary Heart Disease‐Prone Subpopulation.” Texas Heart Institute Journal 31, no. 1: 14–21. https://www.ncbi.nlm.nih.gov/pubmed/15061621. - PMC - PubMed
MeSH terms
Grants and funding
- T32 DE014318/DE/NIDCR NIH HHS/United States
- ZIA HG200373/ImNIH/Intramural NIH HHS/United States
- AAI20042-001/NH/NIH HHS/United States
- 1UM1TR004538/NH/NIH HHS/United States
- R37 AI046326/AI/NIAID NIH HHS/United States
- R21 GM147800/GM/NIGMS NIH HHS/United States
- K23 AG066933/AG/NIA NIH HHS/United States
- ZIA AI001202/ImNIH/Intramural NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- R21GM147800/NH/NIH HHS/United States
- ZIA HG200371/ImNIH/Intramural NIH HHS/United States
- ZIA HG200374/ImNIH/Intramural NIH HHS/United States
- R37AI046326/NH/NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- UM1 TR004538/TR/NCATS NIH HHS/United States
- IP1 CX000875-01A1/U.S. Department of Veterans Affairs
- K23AG066933/NH/NIH HHS/United States
- FA8650-17-2-6816/United States Air Force (DoD)
- Z01 AI000825/ImNIH/Intramural NIH HHS/United States
- IP1 CX000875/CX/CSRD VA/United States
- COVID19-8100-01/U.S. Department of Veterans Affairs
- T32DE014318/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

