Development of an alkaliptosis-related lncRNA risk model and immunotherapy target analysis in lung adenocarcinoma
- PMID: 40264452
- PMCID: PMC12011837
- DOI: 10.3389/fgene.2025.1573480
Development of an alkaliptosis-related lncRNA risk model and immunotherapy target analysis in lung adenocarcinoma
Abstract
Background: Lung cancer has the highest mortality rate among all cancers worldwide. Alkaliptosis is characterized by a pH-dependent form of regulated cell death. In this study, we constructed a model related to alkaliptosis-associated long non-coding RNAs (lncRNAs) and developed a prognosis-related framework, followed by the identification of potential therapeutic drugs.
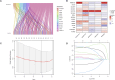
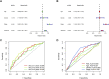
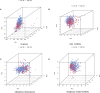
Methods: The TCGA database was utilized to obtain RNA-seq-based transcriptome profiling data, clinical information, and mutation data. We conducted multivariate Cox regression analysis to identify alkaliptosis-related lncRNAs. Subsequently, we employed the training group to construct the prognostic model and utilized the testing group to validate the model's accuracy. Calibration curves were generated to illustrate the discrepancies between predicted and observed outcomes. Principal Component Analysis (PCA) was performed to investigate the distribution of LUAD patients across high- and low-risk groups. Additionally, Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) were conducted. Immune cell infiltration and Tumor Mutational Burden (TMB) analyses were carried out using the CIBERSORT and maftools algorithms. Finally, the "oncoPredict" package was employed to predict immunotherapy sensitivity and to further forecast potential anti-tumor immune drugs. qPCR was used for experimental verification.
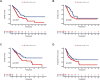
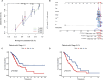
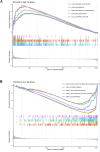
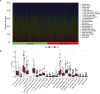
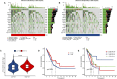
Results: We identified 155 alkaliptosis-related lncRNAs and determined that 5 of these lncRNAs serve as independent prognostic factors. The progression-free survival (PFS) and overall survival (OS) rates of the low-risk group were significantly higher than those of the high-risk group. The risk signature functions as a prognostic factor that is independent of other variables. Different stages (I-II and III-IV) effectively predict the survival rates of lung adenocarcinoma (LUAD) patients, and these lncRNAs can reliably forecast these signatures. GSEA revealed that processes related to chromosome segregation and immune response activation were significantly enriched in both the high- and low-risk groups. The high-risk group exhibited a lower fraction of plasma cells and a higher proportion of activated CD4 memory T cells. Additionally, the OS of the low TMB group was significantly lower compared to the high TMB group. Furthermore, drug sensitivity was significantly greater in the high-risk group than in the low-risk group. These lncRNAs may serve as biomarkers for treating LUAD patients.
Conclusion: In summary, the construction of an alkaliptosis-related lncRNA prognostic model and drug sensitivity analysis in LUAD patients provides new insights into the clinical diagnosis and treatment of advanced LUAD patients.
Keywords: alkaliptosis; immunotherapy sensitivity; lncRNAs; lung adenocarcinoma; prognosis.
Copyright © 2025 Xiong, Liu and Yao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
A methylation-related lncRNA-based prediction model in lung adenocarcinomas.Clin Respir J. 2024 Aug;18(8):e13753. doi: 10.1111/crj.13753. Clin Respir J. 2024. PMID: 39187946 Free PMC article.
-
Cuproptosis-related lncRNA predict prognosis and immune response of lung adenocarcinoma.World J Surg Oncol. 2022 Sep 1;20(1):275. doi: 10.1186/s12957-022-02727-7. World J Surg Oncol. 2022. PMID: 36050740 Free PMC article.
-
A cuproptosis-related lncRNA signature for predicting prognosis and immunotherapy response of lung adenocarcinoma.Hereditas. 2023 Jul 24;160(1):31. doi: 10.1186/s41065-023-00293-w. Hereditas. 2023. PMID: 37482612 Free PMC article.
-
Disulfidptosis-related lncRNAs signature predicting prognosis and immunotherapy effect in lung adenocarcinoma.Aging (Albany NY). 2024 Jun 10;16(11):9972-9989. doi: 10.18632/aging.205911. Epub 2024 Jun 10. Aging (Albany NY). 2024. PMID: 38862217 Free PMC article.
-
Methuosis, Alkaliptosis, and Oxeiptosis and Their Significance in Anticancer Therapy.Cells. 2024 Dec 18;13(24):2095. doi: 10.3390/cells13242095. Cells. 2024. PMID: 39768186 Free PMC article. Review.
References
-
- Capasso M., Brignole C., Lasorsa V. A., Bensa V., Cantalupo S., Sebastiani E., et al. (2024). From the identification of actionable molecular targets to the generation of faithful neuroblastoma patient-derived preclinical models. J. Transl. Med. 22 (1), 151. 10.1186/s12967-024-04954-w - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

