A retrospective analysis comparing metagenomic next-generation sequencing with conventional microbiology testing for the identification of pathogens in patients with severe infections
- PMID: 40264936
- PMCID: PMC12011730
- DOI: 10.3389/fcimb.2025.1530486
A retrospective analysis comparing metagenomic next-generation sequencing with conventional microbiology testing for the identification of pathogens in patients with severe infections
Abstract
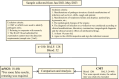
Introduction: The application value of metagenomic next-generation sequencing (mNGS) in detecting pathogenic bacteria was evaluated to promote the rational and accurate use of antibiotics. A total of 180 patients with severe infections were included in this study.
Methods: Based on their different symptoms, bronchoalveolar lavage fluid (BALF) or blood samples were collected for conventional microbiological testing (CMT) and mNGS.
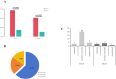
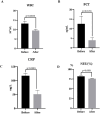
Results: The results indicated that the etiological diagnosis rate of mNGS (78.89%) was significantly higher than that of CMT (20%) (p<0.001). Notably, mNGS exhibited greater sensitivity towards rare pathogens such as Chlamydia pneumoniae, Mycobacterium tuberculosis complex, and Legionella pneumophila, which were undetectable by CMT. Additionally, 64 cases underwent blood culture, BALF culture, and mNGS testing. Analysis revealed that the positive rate of blood culture (3.1%) was lower than that of BALF (25%), and the positive rate of CMT from both types was significantly lower than that of mNGS (89.1%) (p<0.001). In this study, 168 mNGS results were accepted, and 116 patients had their antibiotic therapy adjustment based on mNGS. Paired analysis indicated that white blood cell count (WBC), procalcitonin (PCT), C-reactive protein (CRP), and neutrophil (NEU) percentage provided valuable therapeutic guidance. The survival rate of patients was 55.36%, influenced by patient physical condition and age.
Discussion: Our data indicated that mNGS had significant auxiliary value in the clinical diagnosis and treatment for critically ill patients, especially for those with negative CMT results and clinically undefined infections. mNGS could broaden the detection scope, especially for special pathogens, and improve the detection rate, providing powerful assistance for early clinical diagnosis and treatment.
Keywords: blood; bronchial alveolar lavage fluid; conventional microbiological testing; detection of pathogens; metagenomic next-generation sequencing; severe infections.
Copyright © 2025 Hou, Qiao, Qiao, Shi, Chen, Kong, Hu, Jiang and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
A single-center, retrospective study of hospitalized patients with lower respiratory tract infections: clinical assessment of metagenomic next-generation sequencing and identification of risk factors in patients.Respir Res. 2024 Jun 20;25(1):250. doi: 10.1186/s12931-024-02887-y. Respir Res. 2024. PMID: 38902783 Free PMC article.
-
High utility of bronchoalveolar lavage fluid metagenomic next-generation sequencing approach for etiological diagnosis of pneumonia.BMC Infect Dis. 2024 Nov 2;24(1):1232. doi: 10.1186/s12879-024-10108-6. BMC Infect Dis. 2024. PMID: 39488700 Free PMC article.
-
Clinical application value of simultaneous plasma and bronchoalveolar lavage fluid metagenomic next generation sequencing in patients with pneumonia-derived sepsis.BMC Infect Dis. 2024 Dec 6;24(1):1393. doi: 10.1186/s12879-024-10292-5. BMC Infect Dis. 2024. PMID: 39639243 Free PMC article.
-
Metagenomic next generation sequencing of bronchoalveolar lavage fluids for the identification of pathogens in patients with pulmonary infection: A retrospective study.Diagn Microbiol Infect Dis. 2024 Sep;110(1):116402. doi: 10.1016/j.diagmicrobio.2024.116402. Epub 2024 Jun 12. Diagn Microbiol Infect Dis. 2024. PMID: 38878340 Review.
-
Metagenomic next-generation sequencing on treatment strategies and prognosis of patients with lower respiratory tract infections: A systematic review and meta-analysis.Int J Antimicrob Agents. 2025 Mar;65(3):107440. doi: 10.1016/j.ijantimicag.2024.107440. Epub 2025 Jan 4. Int J Antimicrob Agents. 2025. PMID: 39761759
References
-
- Arvaniti K., Dimopoulos G., Antonelli M., Blot K., Creagh-Brown B., Deschepper M., et al. . (2022). Epidemiology and age-related mortality in critically ill patients with intra-abdominal infection or sepsis: an international cohort study. Int. J. Antimicrobial Agents 60, 106591. doi: 10.1016/j.ijantimicag.2022.106591 - DOI - PubMed
-
- Cai Y. Q., Fang X. Y., Chen Y., Huang Z. D., Zhang C. F., Li W. B., et al. . (2020). Metagenomic next generation sequencing improves diagnosis of prosthetic joint infection by detecting the presence of bacteria in periprosthetic tissues. Int. J. Infect. Dis. 96, 573–578. doi: 10.1016/j.ijid.2020.05.125 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

