Exploring Monogenic, Polygenic, and Epigenetic Models of Common Variable Immunodeficiency
- PMID: 40265101
- PMCID: PMC12014265
- DOI: 10.1155/humu/1725906
Exploring Monogenic, Polygenic, and Epigenetic Models of Common Variable Immunodeficiency
Abstract
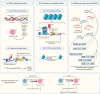
Common variable immunodeficiency (CVID) is the most frequent symptomatic inborn error of immunity (IEI). CVID is genetically heterogeneous and occurs in sporadic or familial forms with different inheritance patterns. Monogenic mutations have been found in a low percentage of patients, and multifactorial or polygenic inheritance may be involved in unsolved patients. In the complex disease model, the epistatic effect of multiple variants in several genes and environmental factors such as infections may contribute. Epigenetic modifications, such as DNA methylation changes, are also proposed to be involved in CVID pathogenesis. In general, the pathogenic mechanism and molecular basis of CVID disease are still unknown, and identifying patterns of association across the genome in polygenic models and epigenetic modification profiles in CVID requires more studies. Here, we describe the current knowledge of the molecular genetic basis of CVID from monogenic, polygenic, and epigenetic aspects.
Keywords: common variable immunodeficiency; etiology; inborn errors of immunity; primary immunodeficiency.
Copyright © 2025 Tayebeh Ranjbarnejad et al. Human Mutation published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

