Multi-Omics Analysis Reveals Differential Molecular Responses of RNA Polymerase Common Subunit ZmRPABC5b for Seedling Development in Maize
- PMID: 40265882
- PMCID: PMC11944614
- DOI: 10.3390/plants14060941
Multi-Omics Analysis Reveals Differential Molecular Responses of RNA Polymerase Common Subunit ZmRPABC5b for Seedling Development in Maize
Abstract
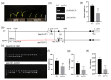
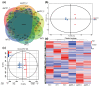
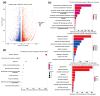
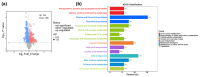
The normal development of maize (Zea mays) seedling is a prerequisite for achieving high crop yields. Although numerous molecular pathways regulate seedling development, the role of RNA polymerases (RNAPs) in this process remains largely unclear, and the function of common RNAP subunits in plants are not well understood. Here, we characterized the loss-of-function mutant of common subunit ZmRPABC5b, defective kernel 701 (dek701), which displays delayed seedling development. To elucidate the role of ZmRPABC5b in maize seedling growth, we conducted transcriptomic and metabolomic analyses. This study found that the loss of ZmRPABC5b function severely impaired early seedling growth, leading to significant reductions in stem length, root length, as well as fresh and dry weight. Transcriptome analysis identified 3780 upregulated and 4385 downregulated differentially expressed genes (DEGs) in dek701 seedlings compared to wild type. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of DEGs revealed that significant enrichment in pathways related to RNA biosynthesis, carbohydrate metabolic, hormone stimulus, cellular transporter and ribosome activity. Metabolome analysis identified 501 differentially expressed metabolites (DEMs) in dek701 seedlings, which were significantly enriched in the amino acid metabolism, secondary metabolites, carbohydrate metabolism, lipid metabolism, transport and translation. These findings provide substantial insight into the ZmRPABC5b regulatory network, positioning it as a central hub for regulating seedling development in maize.
Keywords: RNA polymerase common subunit; maize; metabolome; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

