Genetic deregulation of REL in germinal center B cells induces generation of a pool of lymphoma precursor cells
- PMID: 40267227
- PMCID: PMC12305208
- DOI: 10.1182/bloodadvances.2023012166
Genetic deregulation of REL in germinal center B cells induces generation of a pool of lymphoma precursor cells
Abstract
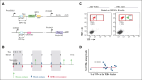
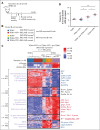
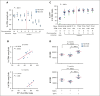
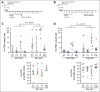
In diffuse large B-cell lymphomas (DLBCLs), gains and amplifications of the 2p15-16 region, which always encompass the REL gene, are mostly restricted to the germinal center (GC) B-cell DLBCL subtype (GCB-DLBCL) for which c-Rel is the pivotal Rel/NF-κΒ subunit. Although REL plays a key role in the GC reaction, its contribution to GCB-DLBCLs remains unclear.To understand the role of REL in the very first steps of GCB transformation, that is, when B cells with deregulated REL are competing with other B cells during chronic antigenic stimulation, we have created a dual-color mouse model that allows to induce REL in a limited pool of activation-induced cytidine deaminase (AID)-imprinted B cells after immunization and to differentially stain AID-imprinted B cells that overexpress REL or not. Dysregulation of REL in AID-imprinted B cells was associated with nuclear c-Rel overexpression in GCs 14 days after immunization. Dysregulation of REL at the GCB stage promoted GCB expansion, which was associated with both class-switch recombination and plasma cell differentiation. REL overexpression conferred a long-term competitive advantage, allowing GC persistence and continuous recirculation of REL-overexpressing B cells. IgHV dominance was increased at the messenger RNA level in REL-overexpressing B cells and clonal expansion was detected at the DNA level in some cases. Highlighting the role of the immune response, our results demonstrate the advantage conferred by REL in the GC competition and provide evidence that, as an oncogenic event of GCBs, its genetic deregulation induces the generation of a long-term pool of lymphoma precursor cells.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




References
-
- Simek S, Rice NR. Detection and characterization of the protein encoded by the chicken c-rel protooncogene. Oncogene Res. 1988;2(2):103–119. - PubMed
-
- Blank V, Kourilsky P, Israël A. NF-kappa B and related proteins: Rel/dorsal homologies meet ankyrin-like repeats. Trends Biochem Sci. 1992;17(4):135–140. - PubMed
-
- Jerkeman M, Hallek M, Dreyling M, Thieblemont C, Kimby E, Staudt L. Targeting of B-cell receptor signalling in B-cell malignancies. J Intern Med. 2017;282(5):415–428. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

