UBL7 is indispensable for spermiogenesis through protecting critical factors from excessive degradation by proteasomes
- PMID: 40268954
- PMCID: PMC12019544
- DOI: 10.1038/s41467-025-59209-z
UBL7 is indispensable for spermiogenesis through protecting critical factors from excessive degradation by proteasomes
Abstract
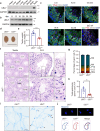
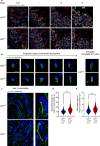
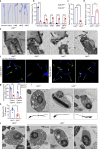
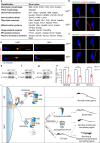
Spermiogenesis is a tightly regulated process to produce mature sperm cells. The ubiquitin-proteasome system (UPS) plays a crucial role in controlling protein half-life and is essential for spermiogenesis. Recently, proteins containing ubiquitin-like domains and ubiquitin-associated domains (UBL-UBA proteins) have emerged as novel regulators within the UPS. In this study, we demonstrate that UBL7, a testis-enriched UBL-UBA protein, is indispensable for sperm formation. Deficiency of UBL7 leads to severe malformations of both the sperm tail and head. Mechanistically, UBL7 interacts with the valosin-containing protein (VCP) complex and proteasomes, and shuttles substrates between them. Notably, UBL7 slows down the degradation rates of substrates involved in endoplasmic reticulum-associated degradation (ERAD) within cells. Through a two-step immunoprecipitation method, we identify several essential factors in spermatids that are protected by UBL7, including factors involved in the development of manchette (such as IFT140), head-tail coupling apparatus (such as SPATA20) and cytoplasmic droplets (such as HK1 and SLC2a3). In summary, our findings highlight UBL7 as a guardian that protects crucial factors from excessive degradation and thereby ensures successful spermiogenesis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Meistrich, M. L. & Hess, R. A. Assessment of spermatogenesis through staging of seminiferous tubules. Spermatogenesis: Methods and Protocols, 299–307 (2013). - PubMed
-
- Dunleavy, J. E., O’Bryan, M. K., Stanton, P. G. & O’Donnell, L. The cytoskeleton in spermatogenesis. Reproduction157, R53–R72 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

