Human de novo mutation rates from a four-generation pedigree reference
- PMID: 40269156
- PMCID: PMC12240836
- DOI: 10.1038/s41586-025-08922-2
Human de novo mutation rates from a four-generation pedigree reference
Abstract
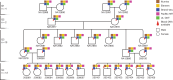
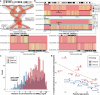
Understanding the human de novo mutation (DNM) rate requires complete sequence information1. Here using five complementary short-read and long-read sequencing technologies, we phased and assembled more than 95% of each diploid human genome in a four-generation, twenty-eight-member family (CEPH 1463). We estimate 98-206 DNMs per transmission, including 74.5 de novo single-nucleotide variants, 7.4 non-tandem repeat indels, 65.3 de novo indels or structural variants originating from tandem repeats, and 4.4 centromeric DNMs. Among male individuals, we find 12.4 de novo Y chromosome events per generation. Short tandem repeats and variable-number tandem repeats are the most mutable, with 32 loci exhibiting recurrent mutation through the generations. We accurately assemble 288 centromeres and six Y chromosomes across the generations and demonstrate that the DNM rate varies by an order of magnitude depending on repeat content, length and sequence identity. We show a strong paternal bias (75-81%) for all forms of germline DNM, yet we estimate that 16% of de novo single-nucleotide variants are postzygotic in origin with no paternal bias, including early germline mosaic mutations. We place all this variation in the context of a high-resolution recombination map (~3.4 kb breakpoint resolution) and find no correlation between meiotic crossover and de novo structural variants. These near-telomere-to-telomere familial genomes provide a truth set to understand the most fundamental processes underlying human genetic variation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: E.E.E. is a scientific advisory board member of Variant Bio. C. Lee is a scientific advisory board member of Nabsys and Genome Insight. D.P. has previously disclosed a patent application (no. EP19169090) relevant to Strand-seq. Z.N.K., C.N., E.D., C.F., C. Lambert, T.M., W.J.R. and M.A.E. are employees and shareholders of PacBio. Z.N.K. is a private shareholder in Phase Genomics. The other authors declare no competing interests.
Figures







References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

