Increased TMEM106B levels lead to lysosomal dysfunction which affects synaptic signaling and neuronal health
- PMID: 40269985
- PMCID: PMC12016085
- DOI: 10.1186/s13024-025-00831-2
Increased TMEM106B levels lead to lysosomal dysfunction which affects synaptic signaling and neuronal health
Abstract
Background: Genetic variation in Transmembrane protein 106B (TMEM106B) is known to influence the risk and presentation in several neurodegenerative diseases and modifies healthy aging. While evidence from human studies suggests that the risk allele is associated with higher levels of TMEM106B, the contribution of elevated levels of TMEM106B to neurodegeneration and aging has not been assessed and it remains unclear how TMEM106B modulates disease risk.
Methods: To study the effect of increased TMEM106B levels, we generated Cre-inducible transgenic mice expressing human wild-type TMEM106B. We evaluated lysosomal and neuronal health using in vitro and in vivo assays including transmission electron microscopy, immunostainings, behavioral testing, electrophysiology, and bulk RNA sequencing.
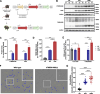
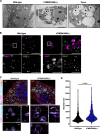
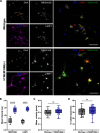
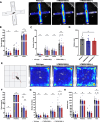
Results: We created the first transgenic mouse model that successfully overexpresses TMEM106B, with a 4- to 8-fold increase in TMEM106B protein levels in heterozygous (hTMEM106B(+)) and homozygous (hTMEM106B(++)) animals, respectively. We showed that the increase in TMEM106B protein levels induced lysosomal dysfunction and age-related downregulation of genes associated with neuronal plasticity, learning, and memory. Increased TMEM106B levels led to altered synaptic signaling in 12-month-old animals which further exhibited an anxiety-like phenotype. Finally, we observed mild neuronal loss in the hippocampus of 21-month-old animals.
Conclusion: Characterization of the first transgenic mouse model that overexpresses TMEM106B suggests that higher levels of TMEM106B negatively impacts brain health by modifying brain aging and impairing the resilience of the brain to the pathomechanisms of neurodegenerative disorders. This novel model will be a valuable tool to study the involvement and contribution of increased TMEM106B levels to aging and will be essential to study the many age-related diseases in which TMEM106B was genetically shown to be a disease- and risk-modifier.
Keywords: Lysosomal dysfunction; Mouse model; Neuronal activity; Synaptic signaling; TMEM106B.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: Dr. Rademakers is a member of the Scientific Advisory Board of Arkuda Therapeutics and receives invention royalties from a patent related to progranulin. Dr. Mackenzie is a member of the Scientific Advisory Board of Prevail Therapeutics and receives invention royalties from a patent related to progranulin. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Doyle JJ, Parker JA, Bateman A. TMEM106B, an unexpected point of contact between FTD, ageing and a hypomyelination disorder. Brain. 2020;143:1628–31. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

