SNORA74A Drives Self-Renewal of Liver Cancer Stem Cells and Hepatocarcinogenesis Through Activation of Notch3 Signaling
- PMID: 40270470
- PMCID: PMC12245033
- DOI: 10.1002/advs.202504054
SNORA74A Drives Self-Renewal of Liver Cancer Stem Cells and Hepatocarcinogenesis Through Activation of Notch3 Signaling
Abstract
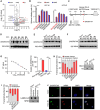
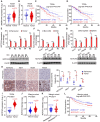
Liver cancer stem cells (CSCs) account for tumor initiation, heterogeneity and therapy resistance. However, the role of small nucleolar RNAs (snoRNAs) in the regulation of liver CSCs remains largely unclear. Here, this work identifies a conserved H/ACA box snoRNA SNORA74A which is highly expressed in liver CSCs. SNORA74A deletion impaired the self-renewal of liver CSCs and suppressed hepatocarcinogenesis. Mechanistically, highly expressed SNORA74A in liver CSCs bound DCAF13 to prevent K48 linked ubiquitination of E2F2 for degradation. E2F2 induced NOTCH3 transcription to initiate Notch3 signaling activation, leading to self-renewal of liver CSCs and hepatocarcinogenesis. Moreover, expression levels of SNORA74A and NOTCH3 are positively related with severity and poor prognosis of hepatocellular carcinoma (HCC) patients. Of note, antisense oligonucleotides (ASOs) against SNORA74A showed effective efficacy for HCC tumors, suggesting SNORA74A might be a potential therapeutic target for HCC therapy by eliminating liver CSCs.
Keywords: DCAF13; SNORA74A; liver cancer stem cells; notch3 signaling; self‐renewal.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
