Disentangling effects of the DR and DQ isomers encoded by the HLA class II haplotype DRB1*15:01/DQB1*06:02 to help establish the true risk allele for FVIII inhibitor development in Hemophilia A
- PMID: 40270541
- PMCID: PMC12016221
- DOI: 10.3389/fgene.2025.1506862
Disentangling effects of the DR and DQ isomers encoded by the HLA class II haplotype DRB1*15:01/DQB1*06:02 to help establish the true risk allele for FVIII inhibitor development in Hemophilia A
Abstract
Introduction: Hemophilia A (HA) patients (HAPs) with the human leukocyte antigen (HLA)-class-II (HLAII) haplotype DRB1*15:01/DQB1*06:02, and thus antigen presenting cells which express HLAII β-polypeptide chains that form heterodimers of DR15- and DQ6-serotypes, respectively, have an increased risk of developing factor (F)VIII inhibitors (FEIs)-neutralizing antibodies against the therapeutic-FVIII-proteins (tFVIIIs) infused to prevent/arrest bleeding. As DRB1*15:01 and DQB1*06:02 exist in strong linkage disequilibrium, association analysis cannot determine which is the actual risk allele.
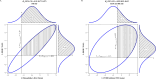
Methods: To establish the true risk allele of this haplotype, we analyzed the tFVIII-derived peptides (tFVIII-dPs) bound to either the DR or DQ molecules that comprise the individual HLAII repertoires expressed by monocyte-derived dendritic cells obtained from 25 normal blood donors and six HAPs, four without and two with FEIs. We performed log-linear mixed model analyses, where the dependent variable is the log of the measured peptide count. Under Model 1, we analyzed an HLAII allele predictor consisting of ten levels (four DRB1 and six DQB1 alleles) in the fixed effects and variables in the random effects to account for non-independence. Model 2-where the HLAII allele variable consisted of only DRB1*15:01 and DQB1*06:02-compares the HLAII alleles.
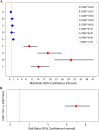
Results: Relative to the Model 1 reference, DRB1*15:01 and DQB1*06:02 significantly increased tFVIII-derived peptide counts, and DRB1*15:01 contributed significantly more than DQB1*06:02. Reported as risk ratios (RRs) and their 95% confidence interval (CI) lower- (LB) and upper-bound (UB), we found a RR (95% CI-LB, -UB) of 14.16 (10.38, 19.33) and 1.76 (1.24, 2.50) for DRB1*15:01 and DQB1*06:02, respectively. Under Model 2, we found an RR for DRB1*15:01 against DQB1*06:02 of 7.00 (5.80, 8.44).
Discussion/conclusion: Our results suggest that DRB1*15:01 is the offending HLAII allele and that DR15 allotypes underlie the increased FEI risk in HAPs.
Keywords: FVIII inhibitors; Hemophilia A; MHC-associated peptide proteomics; dendritic cell protein processing and presentation assays; immunogenic potential; linkage disequilibrium and HLAII haplotype DRB1*15:01/DQB1*06:02; therapeutic FVIII derived peptides; therapeutic FVIII proteins.
Copyright © 2025 Diego, Luu, Almeida, Rajalingam, Hofmann, Galan, Manusov, Powell, Dinh, Mead, Huynh, Verhagen, Peralta, Lehmann, Kumar, Fine, Curran, Goring, Escobar, Williams-Blangero, Maraskovsky, Blangero and Howard.
Conflict of interest statement
These authors have the following positions at Haplogenics Corporation: Director, Technical Operations (BL); Director, Drug Discovery (LD); Chief Medical Officer (JeP); Consultant (HM); and Chief Scientific Officer (TH). Author MH is employed by CSL Innovation GmbH and authors HH, AV, and EM are employed by CSL Limited Research. Author PL is employed by Cellular Technology Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Benjamin R. J., Busch M. P., Fang C. T., Notari E. P., Puren A., Schoub B. D., et al. (2008). Human immunodeficiency virus-1 infection correlates strongly with herpes simplex virus-2 (genital herpes) seropositivity in South African and United States blood donations. Transfusion 48, 295–303. 10.1111/j.1537-2995.2007.01523.x - DOI - PubMed
-
- Diego V. P., Luu B. W., Hofmann M., Dinh L. V., Almeida M., Powell J. S., et al. (2020). Quantitative HLA-class-II/factor VIII (FVIII) peptidomic variation in dendritic cells correlates with the immunogenic potential of therapeutic FVIII proteins in hemophilia A. J. Thrombosis Haemostasis 18 (1), 201–216. 10.1111/jth.14647 - DOI - PubMed
-
- Ettinger R. A., Paz P., James E. A., Gunasekera D., Aswad F., Thompson A. R., et al. (2016). T cells from hemophilia A subjects recognize the same HLA-restricted FVIII epitope with a narrow TCR repertoire. Blood, J. Am. Soc. Hematol. 128 (16), 2043–2054. 10.1182/blood-2015-11-682468 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

