Integration of digital phenotyping, GWAS, and transcriptomic analysis revealed a key gene for bud size in tea plant (Camellia sinensis)
- PMID: 40271457
- PMCID: PMC12015473
- DOI: 10.1093/hr/uhaf051
Integration of digital phenotyping, GWAS, and transcriptomic analysis revealed a key gene for bud size in tea plant (Camellia sinensis)
Abstract
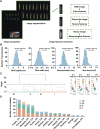
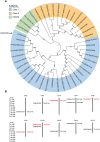
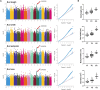
Tea plant (Camellia sinensis) is among the most significant beverage crops globally. The size of tea buds not only directly affects the yield and quality of fresh leaves, but also plays a key role in determining the suitability of different types of tea. Analyzing the genetic regulation mechanism of tea bud size is crucial for enhancing tea cultivars and boosting tea yield. In this study, a digital phenotyping technology was utilized to collected morphological characteristics of the apical buds of 280 tea accessions of representative germplasm at the 'two and a bud' stage. Genetic diversity analysis revealed that the length, width, perimeter, and area of tea buds followed a normal distribution and exhibited considerable variation across natural population of tea plants. Comparative transcriptomic analysis of phenotypic extreme materials revealed a strong negative correlation between the expression levels of four KNOX genes and tea bud size. A key candidate gene, CsKNOX6, was confirmed by further genome-wide association studies (GWAS). Its function was preliminarily characterized by heterologous transformation of Arabidopsis thaliana. Overexpression of CsKNOX6 reduced the leaf area in transgenic plants, which initially determined that it is a key gene negatively regulating bud size. These findings enhance our understanding of the role of KNOX genes in tea plants and provide some references for uncovering the genetic regulatory mechanisms behind tea bud size.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Genome-wide association study of leaf-related traits in tea plant in Guizhou based on genotyping-by-sequencing.BMC Plant Biol. 2023 Apr 12;23(1):196. doi: 10.1186/s12870-023-04192-0. BMC Plant Biol. 2023. PMID: 37046207 Free PMC article.
-
Genome-wide identification of conserved and novel microRNAs in one bud and two tender leaves of tea plant (Camellia sinensis) by small RNA sequencing, microarray-based hybridization and genome survey scaffold sequences.BMC Plant Biol. 2017 Nov 21;17(1):212. doi: 10.1186/s12870-017-1169-1. BMC Plant Biol. 2017. PMID: 29157210 Free PMC article.
-
Differential expression of microRNAs in dormant bud of tea [Camellia sinensis (L.) O. Kuntze].Plant Cell Rep. 2014 Jul;33(7):1053-69. doi: 10.1007/s00299-014-1589-4. Plant Cell Rep. 2014. PMID: 24658841
-
Comparative transcriptomic analysis of the tea plant (Camellia sinensis) reveals key genes involved in pistil deletion.Hereditas. 2020 Sep 8;157(1):39. doi: 10.1186/s41065-020-00153-x. Hereditas. 2020. PMID: 32900387 Free PMC article.
-
Genome-wide characterization of COMT family and regulatory role of CsCOMT19 in melatonin synthesis in Camellia sinensis.BMC Plant Biol. 2024 Jan 16;24(1):51. doi: 10.1186/s12870-023-04702-0. BMC Plant Biol. 2024. PMID: 38225581 Free PMC article.
References
-
- Zhang W, Zhao BT, Yang CM. et al. Research progress on key technology of intelligent picking of high-quality tea. J Chin Agr Mech. 2024;45:202–9
-
- Xu Q, Yang Y, Hu KL. et al. Economic, environmental, and emergy analysis of China's green tea production. Sustain Prod Consum. 2021;28:269–80
-
- Wang HF, Kong FJ, Zhou CE. From genes to networks: the genetic control of leaf development. J Integr Plant Biol. 2021;63:1181–96 - PubMed
LinkOut - more resources
Full Text Sources

